Seed dormancy prevents germination of freshly harvested seeds and is slowly released during seed storage by an unknown mechanism. This work shows that posttranslational modifications of the Arabidopsis thaliana protein DELAY OF GERMINATION1 occurring during seed storage can explain dormancy release.
Abstract
Seed dormancy controls the start of a plant’s life cycle by preventing germination of a viable seed in an unfavorable season. Freshly harvested seeds usually show a high level of dormancy, which is gradually released during dry storage (after-ripening). Abscisic acid (ABA) has been identified as an essential factor for the induction of dormancy, whereas gibberellins (GAs) are required for germination. The molecular mechanisms controlling seed dormancy are not well understood. DELAY OF GERMINATION1 (DOG1) was recently identified as a major regulator of dormancy in Arabidopsis thaliana. Here, we show that the DOG1 protein accumulates during seed maturation and remains stable throughout seed storage and imbibition. The levels of DOG1 protein in freshly harvested seeds highly correlate with dormancy. The DOG1 protein becomes modified during after-ripening, and its levels in stored seeds do not correlate with germination potential. Although ABA levels in dog1 mutants are reduced and GA levels enhanced, we show that DOG1 does not regulate dormancy primarily via changes in hormone levels. We propose that DOG1 protein abundance in freshly harvested seeds acts as a timer for seed dormancy release, which functions largely independent from ABA.
INTRODUCTION
The moment when a seed germinates will determine the environmental conditions that the resulting plant encounters during its life. Accurate timing of seed germination therefore requires a reliable control mechanism. Seed dormancy is a major factor in this control by preventing germination of a viable seed during (temporary) favorable conditions in an unfavorable season (Finch-Savage and Leubner-Metzger, 2006). Low levels of seed dormancy can cause premature germination and seedling mortality. On the contrary, high seed dormancy levels delay germination and decrease the length of the growth season (Donohue et al., 2010). Most crop plants have very low seed dormancy levels, which lead to uniform and fast germination after sowing. However, very low seed dormancy can trigger preharvest sprouting, causing yield losses in cereals (Gubler et al., 2005).
Our knowledge of the molecular regulation of seed dormancy is still incomplete. Based on genetic and physiological studies that were mainly performed in Arabidopsis thaliana, the plant hormone abscisic acid (ABA) has been identified as an essential factor for the induction and maintenance of dormancy. Mutations in genes regulating ABA levels or sensitivity lead to altered seed dormancy levels (Koornneef et al., 1982; Karssen et al., 1983; Nambara and Marion-Poll 2003). Gibberellins (GAs) are required for seed germination, and it is especially the balance between ABA and GA that determines seed dormancy and germination (Finkelstein et al., 2008; Holdsworth et al., 2008). In addition, ethylene can control seed dormancy through its influence on the ABA level (Chiwocha et al., 2005) or ABA signaling (Beaudoin et al., 2000; Ghassemian et al., 2000; Linkies et al., 2009).
Seed dormancy is induced during seed maturation; consequently, mutations in maturation regulators lead to poorly matured seeds with low dormancy levels (Holdsworth et al., 2008). The release of seed dormancy in Arabidopsis requires imbibition at low temperatures (stratification) or dry storage (after-ripening). Several studies reported that stratification involves changes in the levels of and sensitivity to ABA and GA (Ali-Rachedi et al., 2004; Yamauchi et al., 2004), but the precise mechanism of this dormancy release via hormones is still unknown. The release of dormancy by after-ripening is an intriguing process because it occurs in dry seeds with very low humidity levels that prevent active metabolic processes. Nonenzymatic processes have been proposed to alleviate dormancy and experimental evidence for a role of reactive oxygen species in dormancy release by after-ripening in sunflower (Helianthus annuus) has been presented (Oracz et al., 2007; Bazin et al., 2011). An alternative, but not mutually exclusive, theory for the after-ripening mechanism is provided by indirect evidence for the occurrence of humid pockets within dry seeds that would enable transcription and translation of germination factors (Leubner-Metzger, 2005).
Only a few genes regulating dormancy have been identified that are not directly involved in hormone metabolism or seed maturation. Two examples are HISTONE MONOUBIQUITINATION1 (HUB1) and REDUCED DORMANCY2 (RDO2). Their corresponding mutants have been identified in Arabidopsis in a screen for reduced dormancy (Léon-Kloosterziel et al., 1996; Peeters et al., 2002). HUB1 is required for monoubiquitination of histone H2B (Liu et al., 2007), while RDO2 encodes a TFIIS transcription elongation factor (Liu et al., 2011). Both proteins are predicted to interact with the RNA Polymerase II Associated Factor 1 complex, which is involved in chromatin remodeling during transcription elongation. The role of HUB1 and RDO2 in dormancy can largely be explained by their influence on the transcription of other dormancy genes (Liu et al., 2007, 2011).
By contrast, the Arabidopsis gene DELAY OF GERMINATION1 (DOG1) seems to have a more direct role in seed dormancy. DOG1 has been identified as a major quantitative trait locus for seed dormancy in a recombinant inbred line population derived from the lowly dormant accession Landsberg erecta (Ler) and the high dormant accession Cape Verde Islands (Cvi) (Alonso Blanco et al., 2003). DOG1 is a key regulator of seed dormancy; dog1 mutants are completely nondormant and do not show any obvious pleiotropic phenotypes, apart from a reduced seed longevity. DOG1 is alternatively spliced and encodes a protein with unknown molecular function (Bentsink et al., 2006). The DOG1 protein belongs to a small family in Arabidopsis that was recently shown to be conserved in other plant species. DOG1 homologs have been found in the Arabidopsis related species Lepidium sativum and Brassica rapa (Graeber et al., 2010) and in the monocot rice (Oryza sativa; Sugimoto et al., 2010). Interestingly, monocot DOG1 homologs also show functional conservation because ectopic expression of wheat (Triticum aestivum) and barley (Hordeum vulgare) DOG1-like genes induces seed dormancy in Arabidopsis (Ashikawa et al., 2010).
In this study, we reveal a strong correlation between DOG1 protein levels in freshly harvested dry seeds and the time required for after-ripening. The DOG1 protein becomes modified during seed storage, which probably renders it nonfunctional. In addition, we present genetic evidence showing that DOG1 functions independent from ABA. The presence of both DOG1 and ABA is required for seed dormancy. In summary, we propose that DOG1 acts in parallel to ABA signaling and functions as a timer for the release of seed dormancy.
RESULTS
DOG1 mRNA and Protein Levels Show Different Dynamics
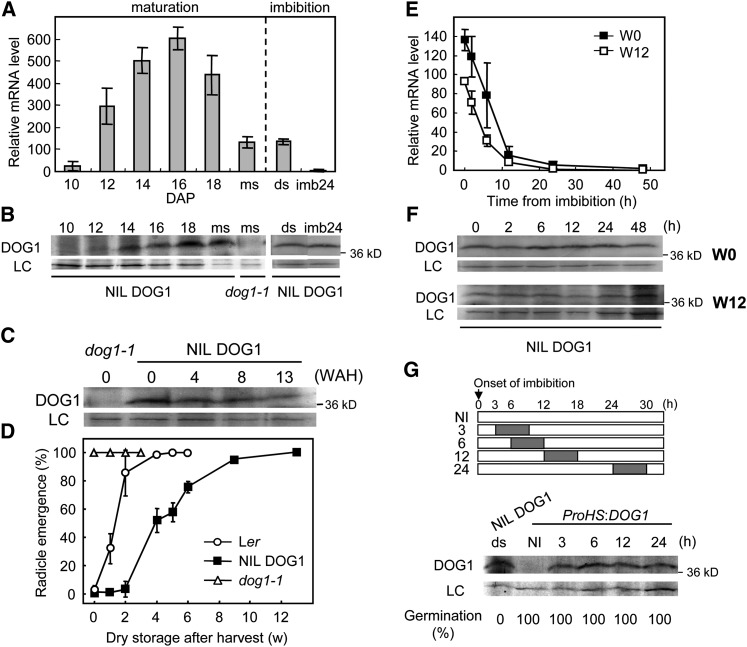
Seed development consists of an embryogenesis phase followed by a seed maturation phase. Seed maturation starts after the embryo has been fully developed and ends when the seed is mature and desiccated, which under our growth conditions occurs at ∼10 d after pollination (DAP) and 20 DAP, respectively. Bentsink et al. (2006) showed by RNA gel blot analysis that DOG1 expression can first be detected at the beginning of seed maturation, peaked around the mid-maturation stage, and decreased toward the end of seed maturation. We confirmed these results by quantitative RT-PCR (qRT-PCR) on siliques and seeds of the highly dormant genotype Near Isogenic Line (NIL) DOG1_Cvi, using primers that amplify all known DOG1 transcript variants from alternative splicing. DOG1 expression shows a peak around 16 DAP, followed by a reduction in expression until ∼20% of the peak level in freshly harvested seeds (Figure 1A). We could also confirm that DOG1 transcript levels quickly disappear after seed imbibition (Figure 1A; Bentsink et al., 2006).
Figure 1.
DOG1 mRNA and Protein Levels Show Different Dynamics during Seed Maturation and Imbibition.
(A) qRT-PCR analysis of DOG1 expression during seed maturation and in freshly harvested and imbibed seeds of NIL DOG1. The DOG1 mRNA level was normalized to the ACT8 mRNA level. ds, freshly harvested dry seeds; imb24, 24 h imbibed seeds; ms, mature seeds (20 DAP). Error bars represent the se of at least three biological replicates.
(B) and (C) DOG1 protein accumulation in NIL DOG1 during seed maturation (B) and in dry seeds during after-ripening (stored at 21°C and ∼50% humidity) (C). The top panel shows DOG1 protein, and the bottom panel a nonspecific band around 60 kD that is used as loading control (LC). The molecular mass marker is shown on the right in kilodaltons. The dog1-1 mutant produces only truncated protein and serves as a negative control. WAH, weeks after harvest.
(D) Germination profiles of the seeds used for the immunoblot analysis in (C). Percentages are means of three biological replicates. The bar represents se.
(E) and (F) DOG1 transcript (E) and protein (F) levels during imbibition in NIL DOG1 freshly harvested (w0) or 12-week dry-stored and fully after-ripened seeds (w12). Transcript levels (E) were analyzed by qRT-PCR, and the values are normalized to ACT8. Error bars represent the se of at least three biological replicates. The immunoblots in (F) for w0 and w12 show DOG1 protein in the top and a loading control in the bottom (LC), similar to (B) and (C).
(G) Top panel shows a schematic representation, indicating the heat treatment given to the transgenic dog1-1 line, containing the DOG1 gene under control of the heat shock promoter (ProHS). The gray bar represents a 6-h heat treatment at 37°C at the indicated time after onset of imbibition. The immunoblots in the bottom panel show the accumulation of DOG1 protein after 6 h of heat induction. ds, dry seeds; NI, noninduced dry seeds. The samples are corresponding to the ones in the top panel. See (B) for details of the immunoblot. The germination percentage of each sample is shown at the bottom.
Similar to DOG1 transcript levels, its protein gradually increased in abundance after the start of seed maturation and reached a peak during the mid-maturation phase. However, in contrast with the transcript level, DOG1 protein did not decrease after reaching its peak but remained at the same level, suggesting that it is a very stable protein (Figure 1B). As a consequence, freshly harvested seeds contain low DOG1 transcript levels but high DOG1 protein levels.
Seed dormancy is released by after-ripening, and we were interested in the dynamics of the DOG1 protein during this process. The amount of DOG1 slightly decreased during dry storage of NIL DOG1 seeds, but was still relatively high after 13 weeks of after-ripening when dormancy had already been fully released (Figures 1C and 1D). We also monitored the dynamics of transcript and protein levels during imbibition using freshly harvested (dormant) and after-ripened seeds. In contrast with DOG1 transcript levels, which quickly disappear after imbibition (Figure 1E), DOG1 protein was hardly affected by imbibition both in dormant and after-ripened seeds (Figure 1F). These results indicate that germination potential is not correlated with DOG1 protein abundance in after-ripened imbibed seeds. Interestingly, induction of DOG1 in imbibed seeds, using a transgenic dog1-1 line containing the DOG1 gene expressed from the heat shock promoter, was also not able to induce dormancy. Seeds from this line germinated 100% when DOG1 was induced by a 6 h 37°C treatment after 3 to 24 h of imbibition (Figure 1G).
DOG1 Expression and Protein Levels in Freshly Harvested Seeds Correlate with Dormancy Levels
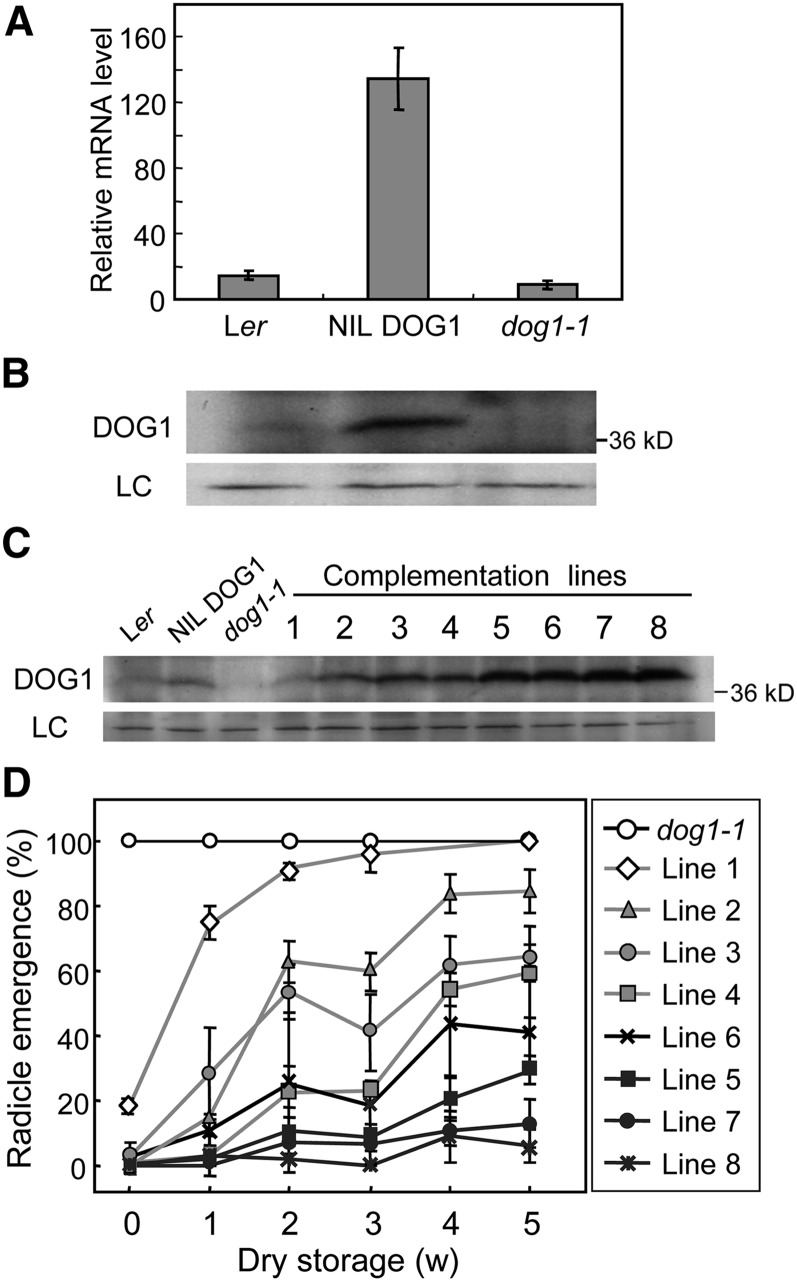
Natural Arabidopsis accessions show considerable differences in seed dormancy, which can for an important part be explained by a quantitative trait locus located at the DOG1 gene, both in laboratory (Bentsink et al., 2010) and field experiments (Huang et al., 2010). In addition, it was shown that differences in DOG1 expression between Arabidopsis accessions contribute to geographical variation in dormancy and germination (Chiang et al., 2011). We confirmed the observation of Bentsink et al. (2006) that the strongly dormant NIL DOG1 genotype shows higher DOG1 transcript levels than the low dormant Ler accession (Figure 2A). In addition, the dog1-1 mutant, which was generated in the NIL DOG1 background, showed strongly decreased DOG1 transcript levels. Consistent with the transcript levels, an immunoblot analysis of seed protein showed that the DOG1 protein level in freshly harvested seeds is more abundant in NIL DOG1 than in Ler (Figure 2B). DOG1 protein could not be detected in the dog1-1 mutant, which does not produce full-length protein because of a one-nucleotide deletion and subsequent frameshift halfway through the gene.
Figure 2.
DOG1 Protein Levels in Freshly Harvested Seeds Highly Correlate with Dormancy Levels.
(A) and (B) DOG1 mRNA (A) and protein levels (B) in mature dry seeds of Ler, NIL DOG1, and dog1-1.
(A) Data from qRT-PCR were normalized to ACT8 mRNA level. Error bars represent the se of at least three biological replicates.
(B) For details of the immunoblot, see Figure 1B.
(C) and (D) DOG1 protein abundance in freshly harvested seeds (C) and germination profiles after dry storage (D) from independent dog1-1 complementation lines, each containing a genomic fragment of DOG1_Ler in a single (or potentially double in line #1) introgression event. Percentages in (D) are means of three biological replicates. The bar represents se.
We further studied the effect of DOG1 protein levels on seed dormancy using eight independent dog1-1 transgenic lines that each contained a genomic fragment of DOG1_Ler in a single introgression event. Analysis of the DOG1 protein levels in freshly harvested seeds from these plants showed a high variation, varying from line 1 with DOG1 protein levels similar to Ler, to lines 7 and 8 in which DOG1 accumulated to much higher levels than NIL DOG1 (Figure 2C). The germination rate during dry seed storage of these lines showed a high correlation with their DOG1 protein levels (Figure 2D). Line 1 showed low dormancy levels and germinated close to 100% after 3 weeks of storage, but lines 7 and 8 germinated <10% after 5 weeks of dry storage.
In summary, we always observed a strong correlation of DOG1 transcript and protein levels in freshly harvested seeds with seed dormancy levels. This indicates that the level of DOG1 protein in freshly harvested seeds determines the level of seed dormancy.
DOG1 Levels Are Upregulated by a Decrease in Temperature during Seed Maturation
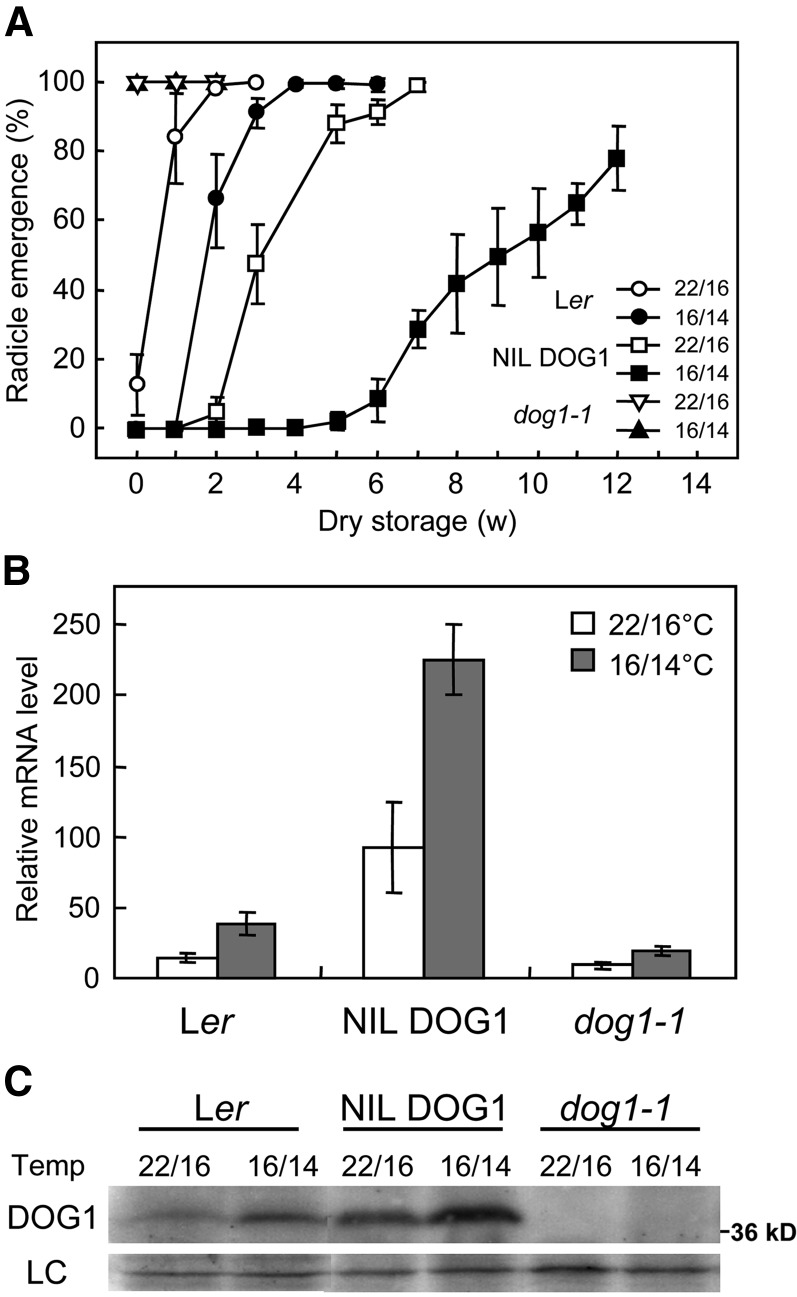
Environmental conditions during seed maturation influence dormancy (Donohue, 2009). We were interested in the role of DOG1 in the dormancy response to temperature during seed maturation. Ler, dog1-1, and NIL DOG1 plants were grown until the onset of flowering under a day/night temperature regime of 22/16°C. Next, half of these plants were transferred to a day/night temperature regime of 16/14°C, while the other half remained at 22/16°C. Seeds subsequently harvested from Ler and NIL DOG1 plants showed increased seed dormancy at the lower seed maturation temperature (Figure 3A).
Figure 3.
Reduced Seed Maturation Temperatures Cause Increased Seed Dormancy and DOG1 Levels.
(A) Germination of Ler, NIL DOG1, and dog1-1 seeds that matured under a day/night regime of 22/16°C or 16/14°C after different periods of dry storage. Harvested seeds were stored at 21°C with ∼50% humidity. Percentages are means of three biological replicates. The bar represents se.
(B) qRT-PCR analysis of DOG1 transcript levels in mature dry seeds. The data were normalized to ACT8. Error bars represent the se of at least three biological replicates.
(C) DOG1 protein levels in mature dry seeds. See Figure 1B for details of the immunoblot. LC, loading control.
Analysis of DOG1 transcript levels in freshly harvested seeds revealed that seeds matured at 16/14°C contained higher DOG1 expression levels compared with seeds that matured at 22/16°C in all three genotypes (Figure 3B). In addition, immunoblot analysis showed enhanced DOG1 protein levels under the cooler seed maturation condition (Figure 3C). This indicated that reduced temperatures during seed maturation enhance DOG1 expression, leading to higher DOG1 protein levels and higher dormancy levels.
DOG1 Protein Changes during After-Ripening
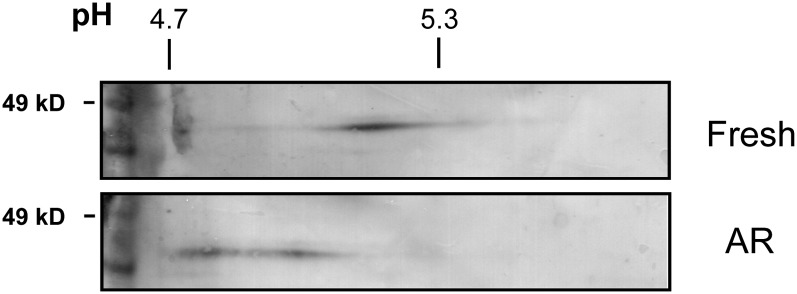
Despite the strong correlation between DOG1 protein levels and dormancy in freshly harvested seeds, such a correlation is completely lacking in after-ripened seeds, suggesting a loss of DOG1 activity during after-ripening. Such a loss of activity could be caused by an altered protein structure. This possibility was investigated by analysis of DOG1 protein on two-dimensional (2D) gels. The DOG1 antibody is not entirely specific and recognizes some additional proteins, which hinders the identification of DOG1 protein on 2D gels. Therefore, we used transgenic Columbia (Col) plants containing 3xHA-tagged DOG1 expressed from the Cvi DOG1 promoter. Figure 4 shows that DOG1 protein of freshly harvested 16 h imbibed seeds is focused at the isoelectric point value of ∼5.15. DOG1 proteins from imbibed seeds after 12 weeks after-ripening were detected at two major spots, which were shifted toward the acidic side. This indicates that the DOG1 protein is altered during the after-ripening process, probably leading to its loss of function. The shift in isoelectric focusing of DOG1 protein during after-ripening has been repeatedly observed in two independent seed batches.
Figure 4.
Altered Isoelectric Focusing of DOG1 Protein during After-Ripening.
Freshly harvested or 12 weeks after-ripened seeds from transgenic plants containing the ProDOG1:3xHA:DOG1 construct were imbibed for 16 h. Extracted proteins were analyzed by 2D gel electrophoresis/immunoblotting using anti-HA antibody. pH values and molecular mass markers are indicated above and on the left of the blots, respectively. AR, after-ripened.
Localization of DOG1 Transcript and Protein
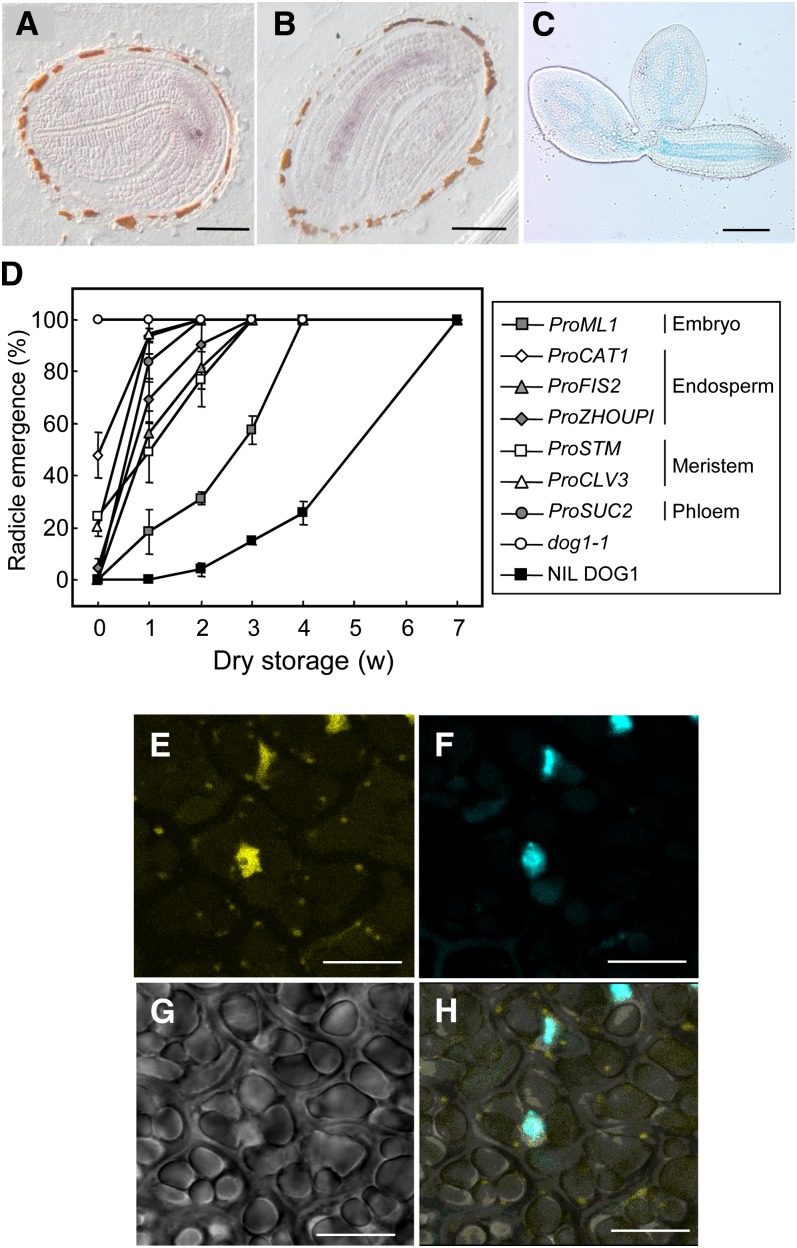
Bentsink et al. (2006) showed that DOG1 expression is seed specific. To determine the tissue-specific distribution of DOG1 expression within the seed, we performed an in situ hybridization analysis using developing seeds during mid-maturation, at which time DOG1 shows its peak expression level. The hybridization signals of the antisense DOG1 probe in consecutive sections were mainly found in vascular tissues of the cotyledon, hypocotyl, and radicle of the embryo (Figures 5A and 5B). Similar experiments using the sense probe did not show any signal. We also monitored the activity of β-glucuronidase (GUS) driven by the DOG1-Cvi promoter. Consistent with the signal from in situ hybridization, GUS activity was observed in vascular tissues (Figure 5C). We conclude that DOG1 is mainly expressed in the vascular tissue of the embryo.
Figure 5.
DOG1 Is Expressed in the Vascular Tissues of Embryos and Is Localized in the Nucleus.
(A) and (B) Spatial patterns of DOG1 mRNA in longitudinal sections through two different seeds at 14 DAP, visualized by in situ hybridization using DOG1 antisense probe. Bars = 100 µm.
(C) DOG1 promoter activity in seeds at 16 to 18 DAP, visualized by GUS activity driven by the 2.6-kb Cvi DOG1 promoter. Embryos were removed from the testa/endosperm after staining with 5-bromo-4-chloro-3-indoryl-β-D-glucuronic acid. Bar = 200 µm.
(D) Germination after different periods of dry storage of dog1-1 and transgenic dog1-1 plants in which DOG1 was expressed from the following tissue-specific promoters: ProSUC2 (phloem), ProSTM and ProCLV3 (apical meristem), ProML1 (epidermal layer of the embryo), and ProZHOUPI, ProCAT1, and ProFIS2 (endosperm). Germination of NIL DOG1 is shown as a dormant control. Percentages are means of three biological replicates. The bar represents se.
(E) to (H) Confocal analysis of the subcellular localization of YFP:DOG1 in embryos at 18 DAP from transgenic dog1-1 plants containing the 2xPro35S:YFP:DOG1 construct. YFP fluorescence (E), 4′,6-diamidino-2-phenylindole staining of nuclei (F), transmission image (G), and merged image (H) of (E) to (G). Bars = 10 µm.
To examine the functional relevance of the tissue-specific expression that we observed, a complementation analysis was performed in which DOG1 was expressed from the following tissue-specific promoters: ProSUC2 (phloem), ProSTM and ProCLV3 (apical meristem), ProML1 (epidermal layer of the embryo), and ProZHOUPI, ProCAT1, and ProFIS2 (endosperm). Interestingly, DOG1 was able to confer some level of dormancy with all of these promoters (Figure 5D). Differences in dormancy level between these misexpression lines could be caused by differences in the amount of produced DOG1 protein and was not further studied. These results suggested that, although DOG1 is mainly expressed from the vascular tissue, it can function independent of where it is expressed in the seed.
To elucidate the subcellular localization of DOG1 protein, we constructed transgenic plants containing the DOG1 genomic fragment with an N-terminal yellow fluorescent protein (YFP) tag that was driven by the 35S promoter and selected lines that complemented the dog1-1 phenotype. The YFP fluorescence was mainly detected in the nucleus (Figures 5E to 5H).
DOG1 Influences the Expression of ABA and GA Metabolic Genes during Imbibition
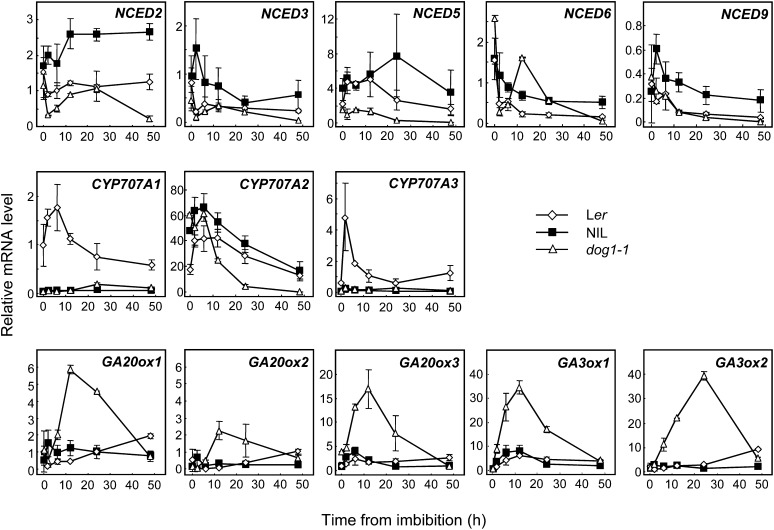
The balance between the ABA and GA pathways determines the dormancy level and germination potential of seeds (Holdsworth et al., 2008). Both dormant and nondormant seeds show a decline in their ABA levels within the first 12 h of imbibition. Thereafter, dormant seeds show consistently higher ABA levels than nondormant seeds (Ali-Rachedi et al., 2004; Lee et al., 2010). We studied the expression kinetics of ABA and GA metabolic genes during imbibition of fresh mature Ler, dog1-1, and NIL DOG1 seeds in detail. Nine-cis-epoxycarotenoid dioxygenase (NCED) genes are key regulatory genes of ABA biosynthesis and were generally upregulated in NIL DOG1 compared with Ler and dog1-1 (Figure 6). The ABA catabolic mutants cyp707a1-3 are defective in ABA 8′-hydroxylase activity and accumulate higher amounts of ABA in dry and imbibed seed (Kushiro et al., 2004; Millar et al., 2006; Okamoto et al., 2006). Among the four genes coding for the enzymes in Arabidopsis, CYP707A2 has been shown to be responsible for degradation of ABA during early imbibition of seeds. Induction of CYP707A1 and CYP707A3 was not observed in NIL DOG1 and dog1-1 (Figure 6). CYP707A2 had similar expression patterns in all three genotypes during imbibition. However, the expression levels in Ler were lower at the beginning of imbibition, while those in dog1-1 were lower at extended imbibition times (Figure 6). Overall, our data suggest that DOG1 is not directly involved in the ABA pathway, but de novo ABA synthesis partially contributes to dormancy maintenance in NIL DOG1.
Figure 6.
Expression of Genes Involved in ABA and GA Metabolism during Imbibition of Freshly Harvested Dormant Ler and NIL DOG1 Seeds and Nondormant dog1-1 Seeds.
Seeds were imbibed and sampled at different time points for RNA extraction. Transcript levels of the key regulatory genes of ABA biosynthesis, NCED2, 3, 5, 6, and 9, the ABA catabolic genes CYP707A1, 2, and 3, and the GA biosynthetic genes GA20ox1, 2, and 3, and GA3ox1 and 2 were analyzed by qRT-PCR. The data were normalized to ACT8 values. Error bars represent the se of at least three biological replicates.
We also examined the expression pattern of several GA biosynthetic genes. The level of GA20ox3 transcript was already higher in dry seeds of dog1-1 compared with NIL DOG1 and Ler. Upon imbibition, GA20ox1, 2, and 3 and GA3ox1 and 2 were all induced in dog1-1 from rather early stages onwards. This induction was not observed in dormant NIL DOG1, suggesting that DOG1 negatively influences GA biosynthesis during imbibition.
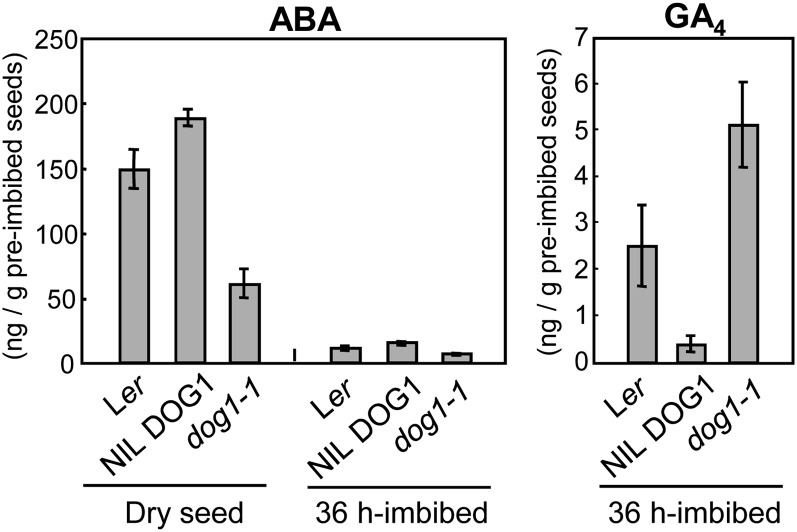
We determined the ABA and GA levels in seeds to find out whether the differential expression of the above-mentioned genes alters the actual hormone levels. Figure 7A shows that the ABA level in fresh mature dry seed was significantly higher in NIL DOG1 compared with the wild type. By contrast, the dog1-1 mutant showed lower ABA levels. We also analyzed 36 h imbibed seeds. As expected, ABA levels were decreased compared with that of dry seeds in all three genotypes. However, they were still higher in NIL DOG1 compared with the dog1-1 mutant (Figure 7). The GA levels in 36 h imbibed seeds showed larger differences between the three genotypes than the ABA levels. GA levels were significantly higher in dog1-1, which was in good accordance with the biosynthetic gene expression pattern (Figure 6). These results indicated that GA production was repressed during imbibition in dormant NIL DOG1 seeds.
Figure 7.
ABA and GA Contents in Dry and Imbibed Seeds of Ler, NIL DOG1, and dog1-1.
ABA and GA were extracted from freshly harvested dry seeds and 36 h imbibed seeds from each genotype. The bar represents se from three independent analyses.
Seed Dormancy Requires Both DOG1 and ABA
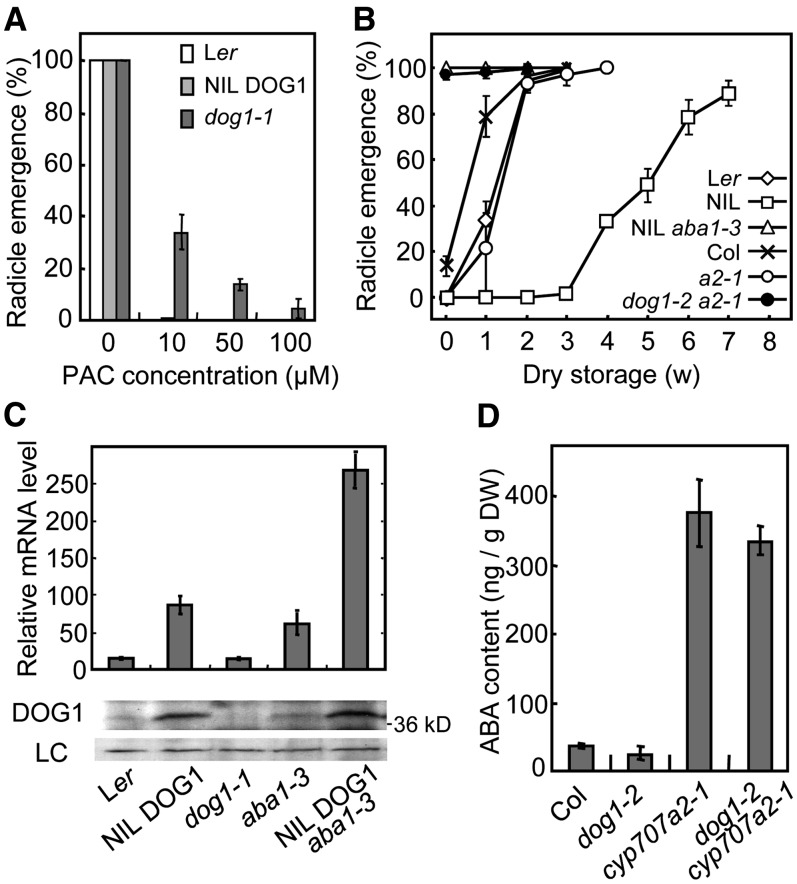
The nondormant phenotype of dog1 mutants is very similar to that of mutants defective in either biosynthesis or signaling of ABA. However, in contrast with those mutants, dog1 cannot germinate without GA as shown by the absence of germination in a double mutant with ga1-3 (Bentsink et al., 2006). In accordance, germination of dog1-1 seeds was sensitive to paclobutrazol, an inhibitor for GA biosynthesis, although at a reduced level compared with NIL DOG1 (Figure 8A). Finally, dog1 has nearly wild-type sensitivity to applied ABA (Bentsink et al., 2006). These data indicated that the nondormant phenotype of dog1-1 was not caused by severe impairment of ABA signaling.
Figure 8.
DOG1 and ABA Are Both Required for the Induction of Seed Dormancy.
(A) Inhibition of germination by the GA biosynthesis inhibitor paclobutrazol on seeds of Ler, NIL DOG1, and dog1-1. Seeds that were after-ripened for 10 weeks were imbibed in the presence of paclobutrazol, and germination percentages were scored. The bars represent se from at least three independent seed batches.
(B) Germination profiles after different periods of dry storage for Ler, NIL DOG1, NIL DOG1 aba1-3, Col, cyp707a2-1, and dog1-2 cyp707a2-1. The cyp707a2-1 mutation is in the Col background. a2-1, cyp707a2-1. Percentages are means of three biological replicates. The bars represent se.
(C) The influence of ABA deficiency on DOG1 expression. The top panel shows DOG1 transcript levels in Ler, NIL DOG1, dog1-1, aba1-3, and NIL DOG1 aba1-3, quantified by qRT-PCR. The values were normalized to the ACT8 level. The bottom panel shows DOG1 protein accumulation of the same genotypes in dry seeds. LC, loading control.
(D) ABA contents in dry seeds of Col, dog1-2, cyp707a2-1, and dog1-2 cyp707a2-1. DW, dry weight.
However, the strong DOG1-Cvi allele is not able to induce dormancy when it is combined with the aba1 mutation (Figure 8B; Bentsink et al., 2006). To determine whether the nondormant phenotype of the aba1 mutant could be explained by reduced DOG1 levels, we checked DOG1 expression in aba1 mutant lines. Surprisingly, our qRT-PCR results showed that DOG1 expression is increased in the aba1 background, regardless of the DOG1 allele (Figure 8C). An immunoblot analysis showed that in accordance with the transcript levels, DOG1 protein levels were higher in ABA-deficient genetic backgrounds (Figure 8C). These data demonstrated that DOG1 function requires ABA and that a feedback regulation exists between these two inducers of dormancy.
Seeds of the cyp707a2 mutant exhibit strongly increased ABA levels and enhanced dormancy (Figures 8B and 8D; Kushiro et al., 2004). We found that seeds from the double mutant dog1-2 cyp707a2-1 show reduced dormancy compared with the cyp707a2-1 single mutant and germinated around 90% directly after harvest (Figure 8B), as reported by Barrero et al. (2010). Analysis of the ABA levels in these mutants indicated strongly increased ABA levels in cyp707a2-1 compared with its wild-type background Col, which was also shown by Kushiro et al. (2004). The dog1-2 cyp707a2-1 double mutant had ABA levels comparable to the cyp707a2-1 single mutant (Figure 8D). This indicated that high ABA accumulation cannot compensate for the absence of DOG1 function. Therefore, DOG1 and ABA are both necessary for the induction of seed dormancy.
DISCUSSION
Seed dormancy has an important function in the plant life cycle and in crop seed management by regulating the timing of germination. However, its intrinsic mechanisms are poorly understood. Thus far, DOG1 is the only identified gene that is absolutely required for seed dormancy without being involved in other processes in the plant. Here, we have shown that DOG1 is a true marker for seed dormancy in Arabidopsis because its protein levels in freshly harvested seeds determine seed dormancy levels.
DOG1 Protein Levels Predict the Dormancy Status of Freshly Harvested Seeds
The amount of DOG1 protein in freshly harvested seeds is highly correlated with the after-ripening time that is required to release seed dormancy. This correlation was observed in seeds with natural DOG1 alleles of different strengths (Figures 2A and 2B) and in transgenic seeds that contain DOG1 alleles varying in expression (Figures 2C and 2D).
The levels of DOG1 protein in ripe seeds are already set at the mid-maturation phase (Figure 1B). The environmental conditions that occur during seed maturation strongly influence dormancy levels (Donohue, 2009). Consistent with this, we have shown that DOG1 protein levels increase by low temperatures during seed maturation, leading to enhanced dormancy (Figure 3C). An increase in seed dormancy levels by lower seed maturation temperatures could be important when Arabidopsis seeds mature in early spring or late autumn in temperate regions. Overall, our work confirms recent observations that DOG1 has an important role in the translation of seed maturation temperature to dormancy levels (Chiang et al., 2011; Footitt et al., 2011; Kendall et al., 2011). It will be of interest to find out whether DOG1 also plays a role in the influence on seed dormancy of other environmental conditions that occur during seed maturation, like light intensity, drought, daylength, and nitrate levels.
DOG1 and ABA Are Both Required for Seed Dormancy but Function in Largely Independent Pathways
The essential roles of the plant hormones ABA and GA in dormancy induction and germination, respectively, are well established (Finkelstein et al., 2008; Holdsworth et al., 2008). It is not surprising that DOG1, being a key regulator of dormancy, influences ABA and GA hormone levels and the transcription levels of genes involved in their metabolism (Figures 6 and 7). However, our data suggest that this is not the direct mode of action for DOG1.
Freshly harvested seeds of the dog1-1 mutant show a strong upregulation of GA biosynthetic genes during the first 24 h of imbibition (Figure 6) and strongly enhanced GA4 levels after 36 h of imbibition compared with NIL DOG1 (Figure 7). This is consistent with the fast and uniform germination of dog1-1 seeds. However, enhanced GA levels are part of the germination process, and reduced biosynthesis of GA is probably an indirect downstream effect of DOG1 function.
In a similar way, DOG1 probably indirectly enhances the biosynthesis of ABA during imbibition. The effect of the different DOG1 alleles on the expression of ABA metabolism genes is not very consistent (Figure 6). However, ABA levels in dry and imbibed seeds are clearly reduced in the dog1-1 mutant and increased in NIL DOG1. Interestingly, ABA and DOG1 have several characteristics in common regarding dormancy. Similar to DOG1, increasing levels of ABA during seed maturation lead to enhanced dormancy levels as demonstrated by ABA-deficient mutants with reduced dormancy (Karssen et al., 1983) and ABA overaccumulators with deeper dormancy (Kushiro et al., 2004; Lefebvre et al., 2006). Furthermore, both ABA and DOG1 are mainly localized in the vascular tissues of the embryo during seed maturation as shown by in situ hybridization (Figures 5A and 5B; Okamoto et al., 2006). Despite these similarities, DOG1 and ABA are very different factors. The plant hormone ABA regulates many aspects of growth, development, and stress responses in plants (Cutler et al., 2010). By contrast, the DOG1 protein is only involved in the regulation of seed dormancy.
Our genetic experiments showed that the presence of both ABA and DOG1 is required to induce seed dormancy. High ABA levels do not lead to dormancy in the absence of DOG1 as shown in the dog1-2 cyp707a2-1 double mutant (Figures 8B and 8D). Similarly, high DOG1 levels cannot compensate for the absence of dormancy in the aba1 mutant (Figure 8B). We found that increased DOG1 protein levels are associated with enhanced ABA levels (Figure 7), whereas the absence of ABA leads to an increase in DOG1 levels (Figure 8C). This indicates the existence of a negative feedback regulation between DOG1 and ABA, which is likely to have a role in fine-tuning of seed dormancy levels. Surprisingly, Kendall et al. (2011) observed reduced DOG1 transcript levels in dry seeds of the aba2-3 mutant and comes to a different conclusion. This might be an allele-specific effect of aba2-3, as we found consistently higher DOG1 levels in the aba1-1 and aba1-3 mutants in different backgrounds (Ler and NIL DOG1). In addition, a microarray analysis of the abi5-7 mutant also showed increased DOG1 expression in dry seeds (Nakabayashi et al., 2005).
Overall, our data suggest that ABA and DOG1 regulate dormancy through largely independent pathways, although they are likely to have downstream targets in common.
Where, How, and When Does DOG1 Regulate Seed Dormancy?
A study of the DOG1 promoter activity and an in situ hybridization analysis indicated that DOG1 is mainly expressed in the vascular tissue of the embryo (Figures 5A to 5C). We could not detect a significant amount of DOG1 transcript in the endosperm. However, microarray data on dissected seeds revealed a low level of DOG1 expression in the endosperm (http://seedgenenetwork.net/Arabidopsis). The vascular tissue connects the meristems at the apical and the radicle side of the embryo. Germination starts with cell elongation in a relatively small zone of the transition zone and lower hypocotyl (Sliwinska et al., 2009), which is later followed by outgrowth of the meristems. Based on the location of its expression, DOG1 might have a role in the inhibition of both processes. Surprisingly, the misexpression experiments showed that DOG1 can induce a low level of dormancy, independent of where it is expressed in the seed. A possible explanation for this unexpected result is that DOG1 (or one of its downstream components) can move through the embryo to its site of action.
DOG1 encodes an unknown protein, and no biochemical function has been reported so far. Transgenic plants containing a YFP:DOG1 fusion protein showed that DOG1 is primarily located in the nucleus, suggesting that DOG1 might act as a transcriptional regulator. DOG1 protein accumulates during seed maturation and remains stable during after-ripening and imbibition. By contrast, DOG1 transcript levels quickly decrease during imbibition (Figure 1), indicating that DOG1 protein will not be produced de novo after reactivation of the translation machinery during germination. However, artificial induction of DOG1 during imbibition using the heat shock promoter did not lead to increased dormancy and could not prevent germination. A possible explanation is that DOG1 is induced in these seeds after the decision to germinate has already been made. Alternatively, DOG1 might be lacking modifications essential for its function in this artificial system.
The amount of DOG1 protein in freshly harvested seeds correlates with the dormancy level of these seeds, but the amount of DOG1 protein in after-ripened seeds does not. We have shown that the DOG1 protein is altered in after-ripened wild-type seeds compared with fresh seeds. This change most likely leads to an altered structure rendering the protein nonfunctional. Therefore, we propose an after-ripening mechanism that consists of a gradual change in DOG1 protein, leading to a decrease in the amount of functional DOG1 in seeds during imbibition. The changes in DOG1 protein during after-ripening are probably not actively driven by enzymes, considering the low moisture content. Oxidative processes by reactive oxygen species would provide an attractive mechanism for the alterations of DOG1 during after-ripening (Oracz et al., 2007).
The above-mentioned observations suggest that DOG1 acts during imbibition of seeds by inhibiting germination. Germination does not require active transcription (Rajjou et al., 2004), and DOG1 might therefore not directly regulate transcription but function by blocking or stimulating translation of stored mRNAs. However, it is likely that DOG1 also acts already during seed maturation because of its effect on ABA levels and transcript levels in dry seeds.
We have shown that DOG1 expression and protein levels in freshly harvested seeds are highly correlated with the dormancy levels of these seeds. DOG1 is a conserved gene that can be found throughout monocots and dicots (Ashikawa et al., 2010; Graeber et al., 2010; Sugimoto et al., 2010). This opens the possibility that DOG1 is a conserved regulator of seed dormancy and that its expression in freshly harvested seeds could be used as a marker to assess or manipulate the seed dormancy level of various crop plant seeds by genetic means.
METHODS
Plant Materials and Growth Conditions
Plant materials used in this study were all derived from the Arabidopsis thaliana accessions Ler, Cvi, and Col-0. Ler and NIL DOG1 have an identical genotype, apart from an ∼4.5 Mb Cvi introgression at the bottom of chromosome 5, containing the DOG1 gene (Alonso-Blanco et al., 2003). The mutant dog1-1 is in the NIL DOG1 background (Bentsink et al., 2006), aba1-3 in the Ler background, and dog1-2 and cyp707a2-1 (Kushiro et al., 2004) in the Col background. The dog1-2 mutant was isolated by V. Raz (Wageningen University and Research Centre, The Netherlands) and identified as a nondormant mutant in the Col background allelic to dog1-1, carrying two nucleotide changes from C to A at the positions 332 and 334 in the first exon, which causes a premature stop codon. The cyp707a2-1 seeds were a gift from Masanori Okamoto (RIKEN). The double homozygous lines of NIL DOG1 aba1-3 and dog1-2 cyp707a2-1 were selected in the F2 progeny of crosses between the two genotypes using PCR to confirm their homozygosity. All plants were sown on soil and grown in a growth chamber with a 16-h-light/8-h-dark cycle (22°C/16°C), unless stated otherwise, or in a greenhouse where the temperature was maintained close to 23°C and 16 h of light was provided daily. Freshly harvested seeds were immediately used for experiments or stored under constant conditions (21°C, 50% humidity, in the dark) for after-ripening treatment.
Germination Tests
About 50 seeds were plated onto a filter paper moistened with demineralized water in Petri dishes and incubated in long-day conditions (16 h light/8 h dark, 25°C/20°C cycle). Radicle emergence was scored after 3 d, since dog1-1 mutant and after-ripened seeds fully germinate within this period. For paclobutrazol responsiveness tests, seeds were stored for 10 weeks to be fully released from dormancy and sown on 0.8% (w/v) water-agarose containing paclobutrazol. Each germination test was done in at least three replicates from independent plants.
RNA Extraction and qRT-PCR
Total RNA was extracted from developing Arabidopsis siliques or imbibed seeds using RNAqueous columns and RNA isolation aid (Ambion) as described previously (Kushiro et al., 2004). For the qRT-PCR, cDNA was synthesized using the QuantiTect reverse transcription kit (Qiagen) from 1 µg of total RNA in a volume of 20 µL, and diluted 20-fold or 200-fold with water for subsequent PCR. qRT-PCR was performed with QuantiTect SYBR Green PCR (Qiagen) on a Mastercycler Realplex2 system (Eppendorf) with the following primer set: DOG1-overall-F, 5′-GAGCTGATCTTGCTCACCGATGTAG-3′; DOG1-overall-R, 5′-CCGCCACCACCTGAAGATTCGTAG-3′. The other primers were described previously; ACTIN8 (ACT8) (Sugliani et al., 2010), NCED2, NCED5, NCED6, NCED9 (Seo et al., 2004), NCED3, CYP707A1, CYP707A2, CYP707A3 (Kushiro et al., 2004), GA20ox1, GA20ox2, GA20ox3, GA3ox1 (Ogawa et al., 2003), and GA3ox2 (Yamauchi et al., 2004). The PCR program was as follows: 15 min at 95°C, followed by 45 cycles of 15 s at 95°C, 20 s at 60°C, and 20 s at 68°C. Single product amplification was validated by melting curve analysis.
The expression value for each gene was quantified using a standard curve with a serial dilution of plasmid of known concentration, and they were normalized to the value of ACT8. At least three biological replicates were analyzed.
Protein Extraction, 2D Gel Electrophoresis, and Immunoblotting
Twenty milligrams of developing siliques or 10 or 5 mg of dry seeds were ground in liquid nitrogen and then extracted with a buffer containing 6 M urea, 2 M thiourea, 0.2% (v/v) Triton X-100, 0.2% (w/v) sarcosyl, and 2 mM DTT in 100 mM Tris-Cl, pH 7.5. After two cycles of 30 min shaking and collecting supernatant by centrifugation at 4°C, protein concentration was determined by Bradford dye reagent (Bio-Rad) using BSA as a standard. Eighty-microgram protein samples were separated by 12% polyacrylamide gel according to Laemmli (1970). Semidry transfer and immunological reaction was performed as previously described (Nakabayashi et al., 1999). DOG1 is detected around 40 kD by immunoblotting despite its calculated molecular mass of around 32 kD. This slow migration is possibly due to the acidity of DOG1 protein, and we confirmed that the DOG1 protein expressed in Escherichia coli shows a similar migration on SDS-PAGE.
For the 2D gel analysis, seed protein was extracted according to Hoehenwarter et al. (2008). Samples were applied to 7-cm immobilized linear pH 4.7 to 5.9 gradient strips (Bio-Rad). Isoelectric focusing was conducted at 250 V for 90 min, gradually increased to 4000 V within 60 min, and held at 4000 V for a total of 20,000 V hours. Proteins were then separated on a 4 to 12% gradient gel (NuPAGE; Invitrogen) and analyzed by immunoblotting using anti-HA monoclonal antibody (HA. 11, clone 16B12; Covance).
Antibody Production
Two oligo peptides C-RRSHGDEDNDNKLRE, corresponding to the 37th to 51st amino acid residues (the 1st C is for coupling site), and GTMRDRRRDCMVDTE, corresponding to the 252nd to 267th residues in DOG1_Cvi, were synthesized, mixed, and used to raise antibodies in rabbits. A small amount of final bleed was affinity purified using each antigen peptide separately (Eurogentec), and the antibody against the first peptide was selected by titer check for further use.
Measurement of Hormone Levels
Extraction, purification, and measurement of ABA were performed as described previously (Yano et al., 2009).
Construction of Transgenic Lines
All the binary constructs were prepared using the Gateway technology (Invitrogen). Binary vectors pGWB1, pGWB3, and pGWB14 (Nakagawa et al., 2007) were gifts from Tsuyoshi Nakagawa (Shimane University, Japan). A 5.46-kb fragment of Ler genomic DNA including 2.51-kb region upstream of the DOG1 start codon, the DOG1 coding region, and 1.2 kb downstream from its stop codon was amplified using the primers 5′-CACCACCAAATTGTTTGTGCATGCTTCAG-3′ and 5′-GACCGGCATTGAAGTCCACA-3′, cloned into pENTR/D-TOPO vector, and converted into pGWB1. The promoter region of 2634 bp from Cvi (2683 to 49 bp upstream of the start codon) was amplified using the primers 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCACCATGAACAAGAACGATTCTC-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCGATCTCTTTTGGTTTGCGTGTTTG-3′, cloned into the pDONR201 vector and converted into pGWB3 for GUS reporter construct. A 2831-bp DOG1 genomic fragment from the starting codon to 3′ downstream from Cvi was amplified using the primers 5′-CACCATGGGATCTTCATCAAAGAA-3′ and 5′-GACCGGCATTGAAGTCCACA-3′, cloned into pENTR/D-TOPO vector and further cloned into 2× Pro35S:YFP binary vector pENSG-YFP for the YFP:DOG1 fusion construct. Underlining in the primer sequences indicates the sequences for the directional cloning into pENTR/D-TOPO vector or the attB sequences for BP reaction (Invitrogen). A 3xHA fragment with NotI adapter was amplified using the following primers (5′-AAGCGGCCGCATGAGCGGGTTAATTAACATCTT-3′ and 5′-AAGCGGCCGCGCTGCACTGAGCAGC-3′; NotI site underlined) from the pGWB14 vector and cloned in the unique NotI site downstream of attL1 sequences in pENTR/D-TOPO vector of above-mentioned entry clone of DOG1_Cvi (ATG to 3′ downstream). The DOG1 promoter from Cvi was cloned in front of the gateway cassette in the pGreen backbone, and the binary construct of the DOG1 promoter_Cvi:3xHA-DOG1_Cvi was produced by LR reaction.
For DOG1 misexpression, the DOG1 genomic sequence was fused with a series of promoters. These were the endosperm-specific promoters Pro At-FIS2, Pro At-ZHOUPI, and Pro Nt-CAT1 (Suzuki et al., 1995; Luo et al., 2000; Yang et al., 2008), the epidermal layer of the embryo-specific promoter Pro At-ML1 (Sessions et al., 1999), the meristem-specific promoters Pro At-STM and Pro At-CLV3 (Long et al., 1996), and the phloem-specific promoter Pro At-SUC2 (Juergensen et al., 2003). At-STM, At-ML1, and At-SUC2 promoter entry clones were a gift from George Coupland (An et al., 2004), and CAT1 promoter was a gift from Gerhard Leubner (Leubner-Metzger, 2005). The At-FIS2, At-ZHOUPI, and At-CLV3 promoters were amplified from Col genomic DNA using specific primers with Gateway tails. All PCR products were introduced into the pDONR207 (Invitrogen) vector through BP reactions, generating promoter entry clones. The specific sequences for each primer pair were pCLV3-F, 5′-CGGATTATCCATAATAAAAACAAA-3′; pCLV3-R, 5′-AGAGAAATATAGAAACTGTTCTTTACT-3′; pFIS2-F, 5′-CTGCGCAGAGAATGAGTACG-3′; pFIS2-R, 5′-GACTGTGATCCACGCAATTTT-3′; pZHOUPI-F, 5′-TTGTGTTACGTTGTAACGAATTTT-3′; and pZHOUPI-R, 5′-TGCTCATTTTACCCTTTTTGC-3′. The DOG1 genomic fragment from Cvi was amplified with primers F (5′-TAAGGTACCTTGACATTTGTCATTGTTT-3′) and R (5′-TAAGGGCCCTTTGGGGTCTAAACCTTGCA-3′) and cloned into a modified pGreen0229 binary vector (An et al., 2004). Underlining in the primer sequences indicates the sequences for the restriction enzymes for the cloning into pGreen0229 vector. Different promoter fusions were produced by LR reactions.
The vector for DOG1-inducible expression was constructed by cloning of the DOG1 genomic sequence into the pLEELA-HSP vector, which was assembled by replacement of the double 35S promoter of the pLEELA vector (which is a derivative of pJawohl3-RNAi; GenBank AF404854) with the promoter from the soybean (Glycine max) Gmhsp 17.6L heat shock protein gene (Severin and Schöffl, 1990).
All the binary constructs were introduced by electroporation into Agrobacterium tumefaciens strain GV3101 or GV3101 carrying the helper plasmid pMP90RK (Koncz and Schell, 1986) or pSoup (Hellens et al., 2000), which were subsequently used to transform dog1-1 mutant or Col plants by floral dipping (Clough and Bent, 1998). All the transgenic lines were first selected based on their antibiotics resistance and further selected by expression level of the transgene or restoration of the mutant phenotype.
RNA in Situ Hybridization
The DOG1 probe, nucleotides 197 to 575 relative to the start codon of the cDNA, was amplified by PCR and used as template for T7-RNA polymerase driven in vitro transcription (Ambion). Siliques of the accession Cvi at 14 DAP were used as samples. Sample preparations and hybridizations were performed as described previously (Coen et al., 1990) with the following modifications. Tween 20 (0.03%) was added to the fixative, and dehydration of the fixed material was done without NaCl. Plant material was embedded in Paraplast+ (Kendall) in an ASP300 tissue processor (Leica). Probes were not hydrolyzed.
Reporter Gene Analysis
For GUS staining, intact siliques or dissected tissues (embryo and testa/endosperm) at 16 to 20 DAP were vacuum infiltrated with 1 mM 5-bromo-4-chloro-3-indoryl-β-D-glucuronic acid solution containing 2 mM potassium ferricyanide and incubated at 37°C overnight (Jefferson, 1987). To terminate the reaction, the tissues were incubated in 70% ethanol and then cleared with chloral hydrate for microscopy. For the analysis of subcellular localization of DOG1, embryos at 16 to 18 DAP were dissected from the testa, and YFP fluorescence was observed using a Leica TCS SP2/AOBS confocal laser scanning microscope.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: DOG1, At5g45830; and ACT8, At1g49240.
Acknowledgments
We thank Christina Philipp for technical assistance, Elmon Schmelzer for help with the confocal microscopy, Matthias Fischer, Anne Harzen, and Jürgen Schmidt for help with the 2D gel analysis, Regina Gentges for the propagation of plant material, and Yusuke Jikumaru, Yumiko Takebayashi, and Yuji Kamiya for the help in hormone analysis. We also thank Vered Raz for the dog1-2 mutant, Ove Nilsson for the heat shock promoter, George Coupland for the At-STM, At-ML1, and At-SUC2 promoter entry clones, Gerd Leubner-Metzger for the Nt-CAT1 promoter, Masanori Okamoto for the cyp707a2-1 seeds, and Tsuyoshi Nakagawa for the vectors pGWB1, pGWB3, and pGWB14. We thank Maarten Koornneef for critical reading of the article. This work was supported by the Max Planck Society, an International Max Planck Research School PhD fellowship (E.M.) and a Deutsche Forschungsgemeinschaft SFB572 grant (W.J.J.S.).
AUTHOR CONTRIBUTIONS
K.N., M.B., and W.J.J.S. designed the research. K.N., M.B., Y.X., and E.M. performed experiments. K.N., R.Y., and M.S. performed hormone measurements. M.B. and S.P. performed the in situ hybridization experiment. K.N., M.B., Y.X., E.M., M.S., and W.J.J.S. analyzed data. K.N. and W.J.J.S. wrote the article.
Glossary
- ABA
abscisic acid
- GA
gibberellin
- Ler
Landsberg erecta
- Ler
Landsberg erecta
- Cvi
Cape Verde Islands
- DAP
days after pollination
- NIL
to be defined
- 2D
two-dimensional
- Col
Columbia
- GUS
β-glucuronidase
- YFP
yellow fluorescent protein
- qRT-PCR
quantitative RT-PCR
References
- Ali-Rachedi S., Bouinot D., Wagner M.H., Bonnet M., Sotta B., Grappin P., Jullien M. (2004). Changes in endogenous abscisic acid levels during dormancy release and maintenance of mature seeds: Studies with the Cape Verde Islands ecotype, the dormant model of Arabidopsis thaliana. Planta 219: 479–488 [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C., Bentsink L., Hanhart C.J., Blankestijn-de Vries H., Koornneef M. (2003). Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics 164: 711–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H., Roussot C., Suárez-López P., Corbesier L., Vincent C., Piñeiro M., Hepworth S., Mouradov A., Justin S., Turnbull C., Coupland G. (2004). CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626 [DOI] [PubMed] [Google Scholar]
- Ashikawa I., Abe F., Nakamura S. (2010). Ectopic expression of wheat and barley DOG1-like genes promotes seed dormancy in Arabidopsis. Plant Sci. 179: 536–542 [DOI] [PubMed] [Google Scholar]
- Barrero J.M., Millar A.A., Griffiths J., Czechowski T., Scheible W.R., Udvardi M., Reid J.B., Ross J.J., Jacobsen J.V., Gubler F. (2010). Gene expression profiling identifies two regulatory genes controlling dormancy and ABA sensitivity in Arabidopsis seeds. Plant J. 61: 611–622 [DOI] [PubMed] [Google Scholar]
- Bazin J., Langlade N., Vincourt P., Arribat S., Balzergue S., El-Maarouf-Bouteau H., Bailly C. (2011). Targeted mRNA oxidation regulates sunflower seed dormancy alleviation during dry after-ripening. Plant Cell 23: 2196–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin N., Serizet C., Gosti F., Giraudat J. (2000). Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 12: 1103–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L., et al. (2010). Natural variation for seed dormancy in Arabidopsis is regulated by additive genetic and molecular pathways. Proc. Natl. Acad. Sci. USA 107: 4264–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L., Jowett J., Hanhart C.J., Koornneef M. (2006). Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 17042–17047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang G.C., Bartsch M., Barua D., Nakabayashi K., Debieu M., Kronholm I., Koornneef M., Soppe W.J.J., Donohue K., De Meaux J. (2011). DOG1 expression is predicted by the seed-maturation environment and contributes to geographical variation in germination in Arabidopsis thaliana. Mol. Ecol. 20: 3336–3349 [DOI] [PubMed] [Google Scholar]
- Chiwocha S.D.S., Cutler A.J., Abrams S.R., Ambrose S.J., Yang J., Ross A.R.S., Kermode A.R. (2005). The etr1-2 mutation in Arabidopsis thaliana affects the abscisic acid, auxin, cytokinin and gibberellin metabolic pathways during maintenance of seed dormancy, moist-chilling and germination. Plant J. 42: 35–48 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coen E.S., Romero J.M., Doyle S., Elliott R., Murphy G., Carpenter R. (1990). floricaula: A homeotic gene required for flower development in Antirrhinum majus. Cell 63: 1311–1322 [DOI] [PubMed] [Google Scholar]
- Cutler S.R., Rodriguez P.L., Finkelstein R.R., Abrams S.R. (2010). Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Donohue K. (2009). Completing the cycle: Maternal effects as the missing link in plant life histories. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364: 1059–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohue K., Rubio de Casas R., Burghardt L., Kovach K., Willis C.G. (2010). Germination, postgermination adaptation, and species ecological ranges. Annu. Rev. Ecol. Evol. Syst. 41: 293–319 [Google Scholar]
- Finch-Savage W.E., Leubner-Metzger G. (2006). Seed dormancy and the control of germination. New Phytol. 171: 501–523 [DOI] [PubMed] [Google Scholar]
- Finkelstein R., Reeves W., Ariizumi T., Steber C. (2008). Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 59: 387–415 [DOI] [PubMed] [Google Scholar]
- Footitt S., Douterelo-Soler I., Clay H., Finch-Savage W.E. (2011). Dormancy cycling in Arabidopsis seeds is controlled by seasonally distinct hormone-signaling pathways. Proc. Natl. Acad. Sci. USA 108: 20236–20241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghassemian M., Nambara E., Cutler S., Kawaide H., Kamiya Y., McCourt P. (2000). Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 12: 1117–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeber K., Linkies A., Müller K., Wunchova A., Rott A., Leubner-Metzger G. (2010). Cross-species approaches to seed dormancy and germination: Conservation and biodiversity of ABA-regulated mechanisms and the Brassicaceae DOG1 genes. Plant Mol. Biol. 73: 67–87 [DOI] [PubMed] [Google Scholar]
- Gubler F., Millar A.A., Jacobsen J.V. (2005). Dormancy release, ABA and pre-harvest sprouting. Curr. Opin. Plant Biol. 8: 183–187 [DOI] [PubMed] [Google Scholar]
- Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hoehenwarter W., van Dongen J.T., Wienkoop S., Steinfath M., Hummel J., Erban A., Sulpice R., Regierer B., Kopka J., Geigenberger P., Weckwerth W. (2008). A rapid approach for phenotype-screening and database independent detection of cSNP/protein polymorphism using mass accuracy precursor alignment. Proteomics 8: 4214–4225 [DOI] [PubMed] [Google Scholar]
- Holdsworth M.J., Bentsink L., Soppe W.J.J. (2008). Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 179: 33–54 [DOI] [PubMed] [Google Scholar]
- Huang X., Schmitt J., Dorn L., Griffith C., Effgen S., Takao S., Koornneef M., Donohue K. (2010). The earliest stages of adaptation in an experimental plant population: Strong selection on QTLS for seed dormancy. Mol. Ecol. 19: 1335–1351 [DOI] [PubMed] [Google Scholar]
- Jefferson R.A. (1987). Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 5: 387–405 [Google Scholar]
- Juergensen K., Scholz-Starke J., Sauer N., Hess P., van Bel A.J., Grundler F.M. (2003). The companion cell-specific Arabidopsis disaccharide carrier AtSUC2 is expressed in nematode-induced syncytia. Plant Physiol. 131: 61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karssen C.M., Brinkhorst Van der Swan D.L.C., Breekland A.E., Koornneef M. (1983). Induction of dormancy during seed development by endogenous abscisic acid - studies on abscisic-acid deficient genotypes of Arabidopsis thaliana (L) Heynh. Planta 157: 158–165 [DOI] [PubMed] [Google Scholar]
- Kendall S.L., Hellwege A., Marriot P., Whalley C., Graham I.A., Penfield S. (2011). Induction of dormancy in Arabidopsis summer annuals requires parallel regulation of DOG1 and hormone metabolism by low temperature and CBF transcription factors. Plant Cell 23: 2568–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C., Schell J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204: 383–396 [Google Scholar]
- Koornneef M., Jorna M.L., Brinkhorst Van der Swan D.L.C., Karssen C.M. (1982). The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L). Heynh. Theor. Appl. Genet. 61: 385–393 [DOI] [PubMed] [Google Scholar]
- Kushiro T., Okamoto M., Nakabayashi K., Yamagishi K., Kitamura S., Asami T., Hirai N., Koshiba T., Kamiya Y., Nambara E. (2004). The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: Key enzymes in ABA catabolism. EMBO J. 23: 1647–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lee K.P., Piskurewicz U., Turecková V., Strnad M., Lopez-Molina L. (2010). A seed coat bedding assay shows that RGL2-dependent release of abscisic acid by the endosperm controls embryo growth in Arabidopsis dormant seeds. Proc. Natl. Acad. Sci. USA 107: 19108–19113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V., North H., Frey A., Sotta B., Seo M., Okamoto M., Nambara E., Marion-Poll A. (2006). Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 45: 309–319 [DOI] [PubMed] [Google Scholar]
- Léon-Kloosterziel K.M., van de Bunt G.A., Zeevaart J.A.D., Koornneef M. (1996). Arabidopsis mutants with a reduced seed dormancy. Plant Physiol. 110: 233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leubner-Metzger G. (2005). Beta-1,3-glucanase gene expression in low-hydrated seeds as a mechanism for dormancy release during tobacco after-ripening. Plant J. 41: 133–145 [DOI] [PubMed] [Google Scholar]
- Linkies A., Müller K., Morris K., Turecková V., Wenk M., Cadman C.S.C., Corbineau F., Strnad M., Lynn J.R., Finch-Savage W.E., Leubner-Metzger G. (2009). Ethylene interacts with abscisic acid to regulate endosperm rupture during germination: A comparative approach using Lepidium sativum and Arabidopsis thaliana. Plant Cell 21: 3803–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Geyer R., van Zanten M., Carles A., Li Y., Hörold A., van Nocker S., Soppe W.J.J. (2011). Identification of the Arabidopsis REDUCED DORMANCY 2 gene uncovers a role for the polymerase associated factor 1 complex in seed dormancy. PLoS ONE 6: e22241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Koornneef M., Soppe W.J.J. (2007). The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell 19: 433–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J.A., Moan E.I., Medford J.I., Barton M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Luo M., Bilodeau P., Dennis E.S., Peacock W.J., Chaudhury A. (2000). Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 97: 10637–10642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A.A., Jacobsen J.V., Ross J.J., Helliwell C.A., Poole A.T., Scofield G., Reid J.B., Gubler F. (2006). Seed dormancy and ABA metabolism in Arabidopsis and barley: The role of ABA 8′-hydroxylase. Plant J. 45: 942–954 [DOI] [PubMed] [Google Scholar]
- Nakabayashi K., Ito M., Kiyosue T., Shinozaki K., Watanabe A. (1999). Identification of clp genes expressed in senescing Arabidopsis leaves. Plant Cell Physiol. 40: 504–514 [DOI] [PubMed] [Google Scholar]
- Nakabayashi K., Okamoto M., Koshiba T., Kamiya Y., Nambara E. (2005). Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: Epigenetic and genetic regulation of transcription in seed. Plant J. 41: 697–709 [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nambara E., Marion-Poll A. (2003). ABA action and interactions in seeds. Trends Plant Sci. 8: 213–217 [DOI] [PubMed] [Google Scholar]
- Ogawa M., Hanada A., Yamauchi Y., Kuwahara A., Kamiya Y., Yamaguchi S. (2003). Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15: 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M., Kuwahara A., Seo M., Kushiro T., Asami T., Hirai N., Kamiya Y., Koshiba T., Nambara E. (2006). CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol. 141: 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oracz K., El-Maarouf Bouteau H., Farrant J.M., Cooper K., Belghazi M., Job C., Job D., Corbineau F., Bailly C. (2007). ROS production and protein oxidation as a novel mechanism for seed dormancy alleviation. Plant J. 50: 452–465 [DOI] [PubMed] [Google Scholar]
- Peeters A.J.M., Blankestijn-De Vries H., Hanhart C.J., Léon-Kloosterziel K.M., Zeevaart J.A.D., Koornneef M. (2002). Characterization of mutants with reduced seed dormancy at two novel rdo loci and a further characterization of rdo1 and rdo2 in Arabidopsis. Physiol. Plant. 115: 604–612 [DOI] [PubMed] [Google Scholar]
- Rajjou L., Gallardo K., Debeaujon I., Vandekerckhove J., Job C., Job D. (2004). The effect of alpha-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol. 134: 1598–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M., Aoki H., Koiwai H., Kamiya Y., Nambara E., Koshiba T. (2004). Comparative studies on the Arabidopsis aldehyde oxidase (AAO) gene family revealed a major role of AAO3 in ABA biosynthesis in seeds. Plant Cell Physiol. 45: 1694–1703 [DOI] [PubMed] [Google Scholar]
- Sessions A., Weigel D., Yanofsky M.F. (1999). The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 20: 259–263 [DOI] [PubMed] [Google Scholar]
- Severin K., Schöffl F. (1990). Heat-inducible hygromycin resistance in transgenic tobacco. Plant Mol. Biol. 15: 827–833 [DOI] [PubMed] [Google Scholar]
- Sliwinska E., Bassel G.W., Bewley J.D. (2009). Germination of Arabidopsis thaliana seeds is not completed as a result of elongation of the radicle but of the adjacent transition zone and lower hypocotyl. J. Exp. Bot. 60: 3587–3594 [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Takeuchi Y., Ebana K., Miyao A., Hirochika H., Hara N., Ishiyama K., Kobayashi M., Ban Y., Hattori T., Yano M. (2010). Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc. Natl. Acad. Sci. USA 107: 5792–5797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugliani M., Brambilla V., Clerkx E.J.M., Koornneef M., Soppe W.J.J. (2010). The conserved splicing factor SUA controls alternative splicing of the developmental regulator ABI3 in Arabidopsis. Plant Cell 22: 1936–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Miyamoto R., Hattori T., Nakamura K., Asahi T. (1995). Differential regulation of the expression in transgenic tobacco of the gene for beta-glucuronidase under the control of the 5′-upstream regions of two catalase genes from castor bean. Plant Cell Physiol. 36: 273–279 [DOI] [PubMed] [Google Scholar]
- Yamauchi Y., Ogawa M., Kuwahara A., Hanada A., Kamiya Y., Yamaguchi S. (2004). Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 16: 367–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Johnston N., Talideh E., Mitchell S., Jeffree C., Goodrich J., Ingram G. (2008). The endosperm-specific ZHOUPI gene of Arabidopsis thaliana regulates endosperm breakdown and embryonic epidermal development. Development 135: 3501–3509 [DOI] [PubMed] [Google Scholar]
- Yano R., Kanno Y., Jikumaru Y., Nakabayashi K., Kamiya Y., Nambara E. (2009). CHOTTO1, a putative double APETALA2 repeat transcription factor, is involved in abscisic acid-mediated repression of gibberellin biosynthesis during seed germination in Arabidopsis. Plant Physiol. 151: 641–654 [DOI] [PMC free article] [PubMed] [Google Scholar]