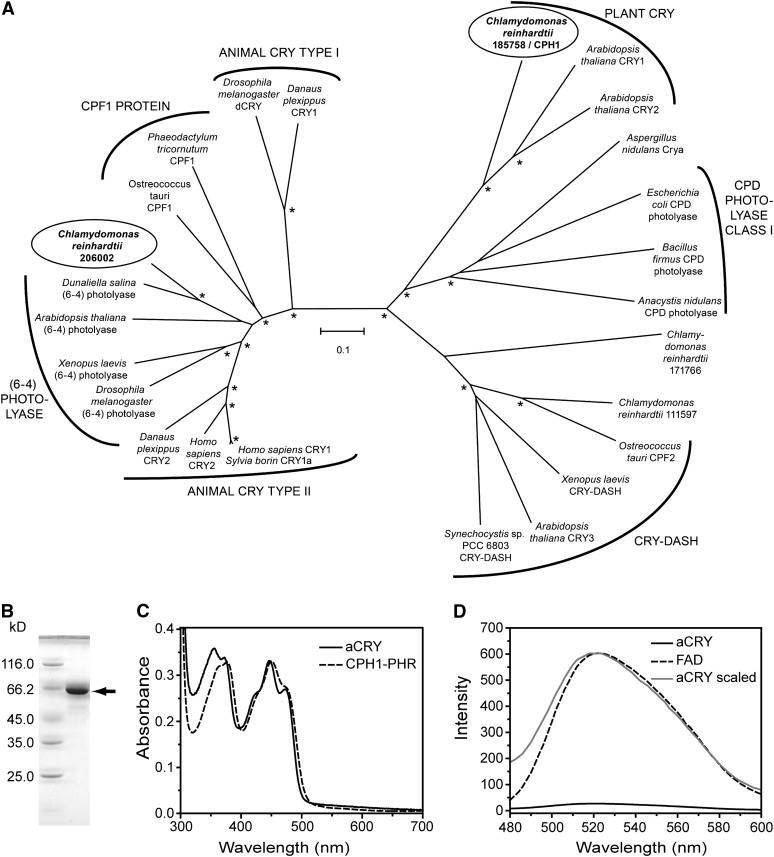
Figure 1.
Phylogenetic Position of aCRY and Characterization of Heterologously Expressed aCRY.
(A) aCRY from C. reinhardtii (gene 206002; circled) groups with type II animal CRYs, (6-4) photolyases, and CPF1 proteins but not with the CPH1 plant CRY from C. reinhardtii (gene 185758; circled). For simplicity, photolyases other than members of cyclobutane pyrimidine dimer (CPD) class I and (6-4) were omitted. Bootstrap values larger than 95% are indicated by asterisks. Numbering of uncharacterized sequences from C. reinhardtii is according to Joint Genome Institute Chlre version 4.0. The scale bar indicates amino acid substitutions per site.
(B) SDS-PAGE of heterologously expressed, full-length aCRY with a His tag. A single band is detected and marked with an arrow. This band migrates to a position that aligns with the marker protein BSA at a molecular mass of 66.2 kD.
(C) UV/Vis spectrum of purified aCRY. The absorption spectrum shows the characteristic pattern of protein-bound, oxidized flavin with absorbance maxima at ∼360 and 447 nm. The spectrum differs from that of the PHR domain of CPH1, the plant CRY of C. reinhardtii (dashed line), which suggests a different hydrogen bonding environment of the flavin.
(D) Fluorescence emission spectrum of aCRY excited at 447 nm (solid line) as compared with that of free FAD (dashed line). The weak emission of FAD is further reduced by binding to aCRY. A scaled trace of aCRY (gray) demonstrates the shift in the fluorescence maximum of free FAD by 2 nm in native aCRY.