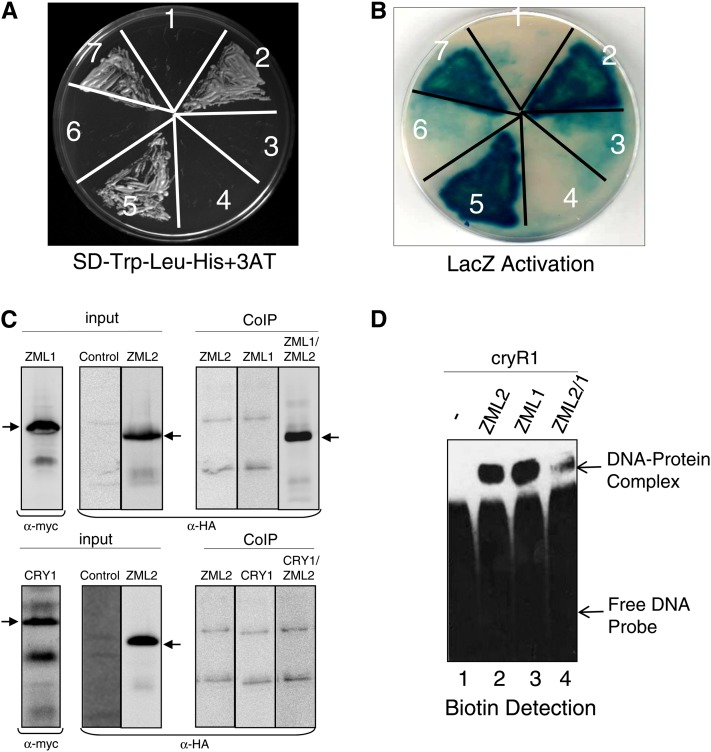
Figure 8.
Heterodimerization of ZML2 and ZML1.
(A) and (B) The C-terminal domain of CRY1 protein does not interact with ZML2 and ZML1 proteins (3 and 6), whereas ZML2 and ZML1 homo- and heterodimerize in the yeast two-hybrid system as indicated by the activation of HIS3 reporter gene (A) and by the LacZ activation (B). Growth due to the activation of HIS3 reporter gene was examined in the presence of 3-AT. NMY51 yeast strain was cotransformed with ZML2 bait and empty prey vector (negative control) (1), ZML2 bait and ZML1 prey (2), ZML2 bait and CRY1-CT prey (3), ZML1 bait and empty prey vector (negative control) (4), ZML1 bait and ZML1 prey (5), ZML1 bait and CRY1-CT prey (6), and p53 bait and Large T prey (positive control) (7).
(C) Interaction was observed between ZML2 and ZML1, but not ZML2 and CRY1, using an in vivo Co-IP assay. HA-ZML2, cMYC-ZML1, and cMYC-CRY1 fusion proteins were expressed in Arabidopsis protoplasts, and Co-IP was performed using anti-cMYC monoclonal antibody bound to protein G–coated magnetic beads. Input and Co-IP fractions were detected by immunoblot analysis with anti-cMYC and HRP-conjugated anti-HA monoclonal antibodies, respectively. The arrows indicate the position of the respective fusion proteins.
(D) EMSA testing DNA binding activity of ZML1 and heterodimerization with ZML2. Signals were detected using a chemiluminescent nucleic acid detection module. Lane 1, free DNA probe; lane 2, ZML2 protein incubated with DNA probe; lane 3, ZML1 protein incubated with DNA probe; lane 4, ZML2 and ZML1 proteins with DNA probe. The migration positions of the free DNA probe (bottom band) and the DNA-protein complexes (top band) are marked by arrows.
[See online article for color version of this figure.]