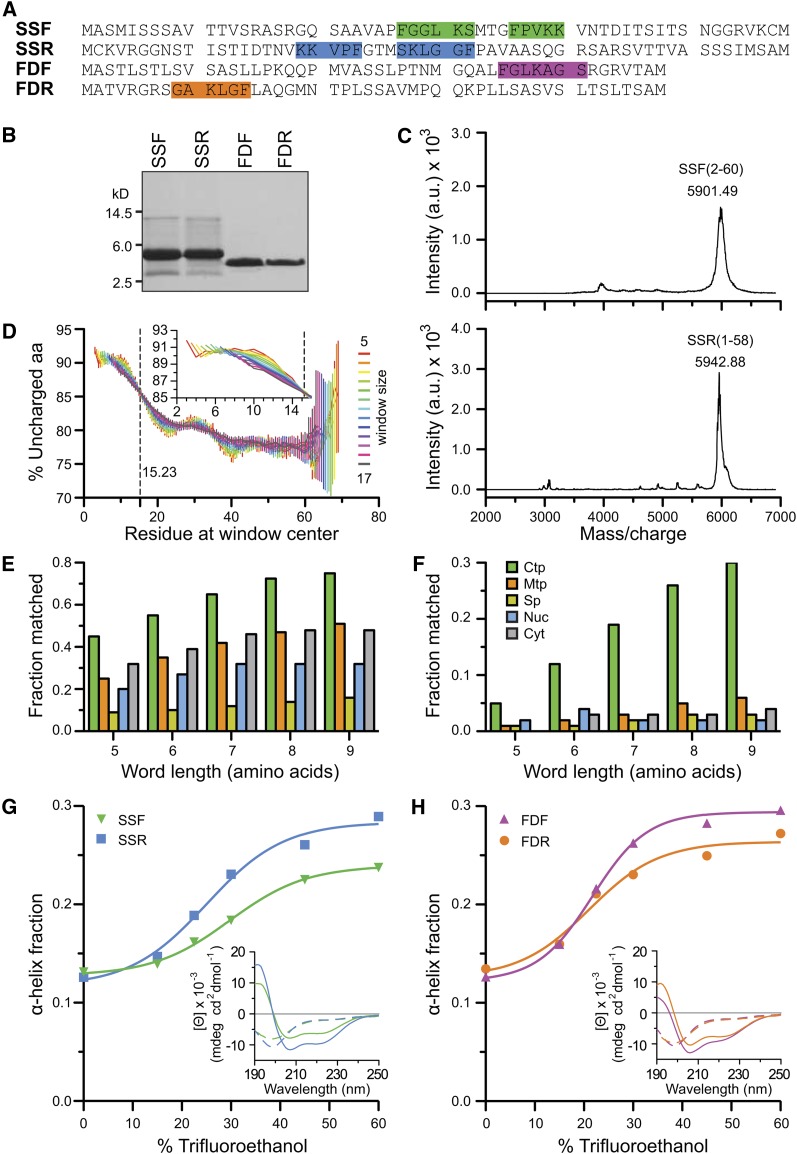
Figure 1.
Purification of Forward and Retro-TPs and Bioinformatic and Secondary Structure Analyses of TPs.
(A) The sequences of SSF, SSR, FDF, and FDR. FGLK motifs are highlighted.
(B) SDS-PAGE gel of the four peptides.
(C) MALDI-TOF spectra of SSF and SSR peptides. a.u., arbitrary units.
(D) Analysis of N-terminal uncharged region in TP data set (n = 912). Means ± se are shown. Inset shows zoom-in without error bars. aa, amino acids.
(E) and (F) The percentage of sequences matched to the TargetP results using a flexible rule 35 and a strict rule 22, respectively. The rules are defined in Supplemental Table 3 online. Ctp, predicted chloroplast localization; Cyt, predicted cytoplasm; Mtp, predicted mitochondrial localization; Nuc, predicted nuclear localization; Sp, predicted secretory pathway.
(G) and (H) Both forward and reverse peptides share similar TFE-induced random coil to α-helix transitions. Insets report the representative CD spectra. Dashed and solid lines generated from 0 and 60% TFE, respectively.