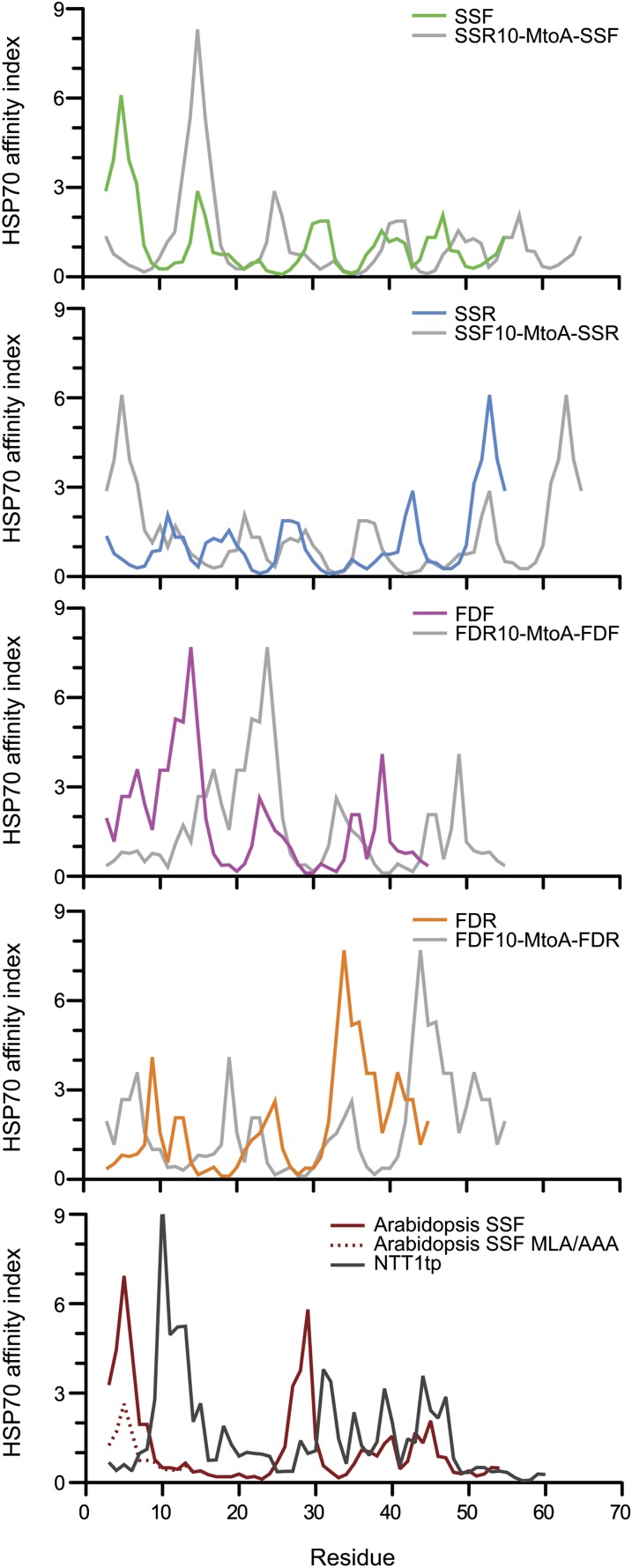
Figure 7.
Hsp70 Binding Site Prediction.
Two Hsp70 binding site prediction algorithms developed from E. coli Hsp70, DnaK, and binding assays were used in the analysis. Prediction results using algorithms by Ivey et al. (2000) derived from Gragerov et al. (1994) are shown here, and those of Rüdiger et al. (1997) are shown in Supplemental Figure 3 online. The import-efficient peptides seem to harbor a strong predicted Hsp70 binding site at the N terminus (area under curve within the first 10 amino acids) where it is lacking in the import-deficient peptides. The results generated from the algorithm derived from Gragerov et al. show strong agreement in every prediction. However, the algorithm developed by Rüdiger et al. show some disagreement in the predictions of SSF and SSR where the calculated energy contribution within the first 10 residues shows no difference. NTT1tp is the TP of Arabidopsis nucleotide transporter 1.