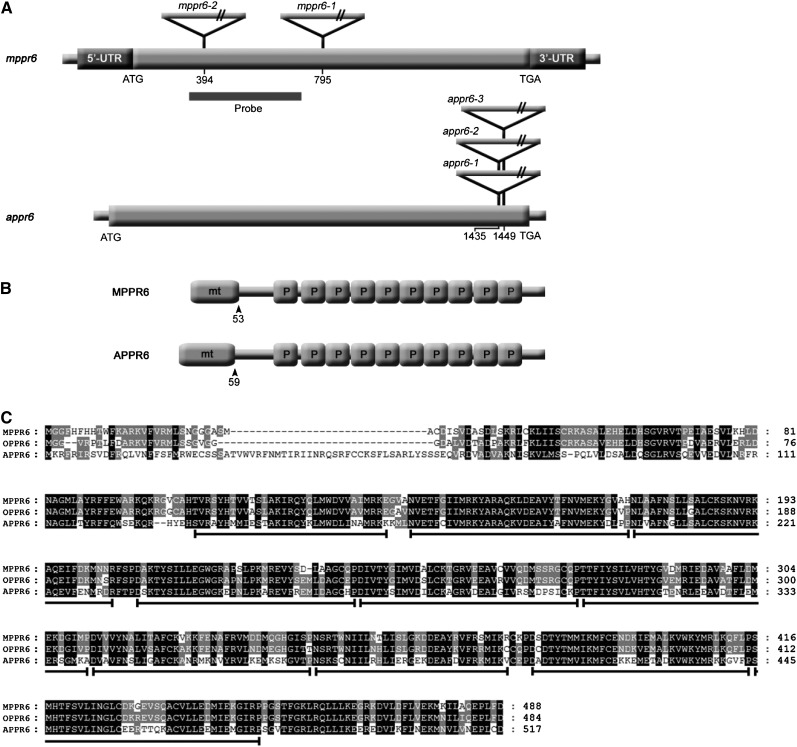
Figure 4.
cDNA and Protein Structure of MPPR6 and APPR6.
(A) Schematic representation of mppr6 and appr6 cDNAs (diagrams not to scale). Mu (maize) and T-DNA insertions (Arabidopsis) are indicated by triangles. Positions of insertions with respect to the start codon (ATG) are shown in both cDNAs. ORFs are represented by wide gray bars, whereas dark-gray bars indicate the 5′ and 3′ UTRs in mppr6 cDNA.
(B) Protein motif organization of MPPR6 and APPR6. Mitochondrial targeting signals (mt) and PPR motifs (P) are indicated. Amino acid positions of transit peptide cleavage sites are shown by arrowheads.
(C) Alignment of MPPR6 (maize), OPPR6 (rice), and APPR6 (Arabidopsis) orthologs. The homologous areas are highlighted. PPR motifs in the sequences are indicated below by black lines.