Summary
The Hippo signaling pathway emerged from studies of Drosophila tumor suppressor genes, and is now appreciated as a major growth control pathway in vertebrates as well as arthropods. As a recently discovered pathway, key components of the pathway are continually being identified, and new insights into how the pathway is regulated and deployed are arising at a rapid pace. Over the past year and a half, significant advances have been made in our understanding of upstream regulatory inputs into Hippo signaling, key negative regulators of Hippo pathway activity have been identified, and important roles for the pathway in regeneration have been described. This review describes these and other advances, focusing on recent progress in our understanding of Hippo signaling that has come from continued studies in Drosophila.
Introduction
The formation of complex and varied structures like eyes, arms, heart and brain from an egg during development raises many intriguing questions. One that has long engaged developmental biologists is: what mechanisms are used to regulate growth to form organs of a particular size? A related question is how this size is remembered throughout the life of an organism, to regulate homeostasis and regeneration. For example, when two-thirds of a rat’s liver is removed, the cells in the remaining third proliferate to restore the organ to its normal size (Michalopoulos and DeFrances, 1997). What mechanisms are used to tell the proliferating cells that the liver has grown back to its original size and that it is time to exit the cell cycle? The discovery of the Hippo signaling pathway and its crucial role in growth regulation has begun to suggest answers to these questions, and to provide a better understanding of the molecular mechanisms controlling organ size during development and regeneration (reviewed in Reddy and Irvine, 2008; Kango-Singh and Singh, 2009; Pan, 2010; Zhao et al., 2010; Halder and Johnson, 2011).
As for a number of other intercellular signaling pathways, many of the key components and basic signal transduction steps associated with Hippo signaling were first discovered in Drosophila, and later shown to be conserved among other animals. Progress in understanding Hippo signaling recently has come at a remarkable pace, and many new and exciting discoveries are now coming from studies on vertebrates and mammalian cell culture models. Nonetheless, studies in Drosophila continue to provide a crucial foundation for further advances in our understanding and appreciation of Hippo signaling. Here, we focus this review on the Drosophila Hippo pathway, highlighting some of the most recent discoveries and their implications for our understanding of the role and regulation of Hippo signaling in controlling organ size.
The Hippo kinase cassette
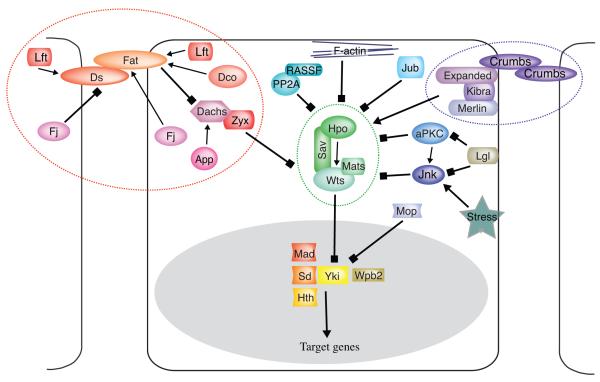
At the core of the Hippo pathway are four proteins, Hippo (Hpo), Salvador (Sav), Warts (Wts), and Mob-as-tumor-suppressor (Mats), which together form the Hippo kinase cassette (Fig 1, Table 1). They were all first identified through genetic screens as Drosophila tumor suppressors (Justice et al., 1995; Xu et al., 1995; Kango-Singh et al., 2002; Tapon et al., 2002; Harvey et al., 2003; Jia et al., 2003; Pantalacci et al., 2003; Udan et al., 2003; Wu et al., 2003; Lai et al., 2005). Loss-of-function mutations in any of these four genes result in overgrowth due to increased cell proliferation and growth, and reduced cell death (Fig. 2A). Hpo and Wts are both Ser/Thr kinases that are activated by phosphorylation, and act sequentially within the pathway. Biochemical studies of these proteins, together with studies of their mammalian homologues, have indicated that Hpo activation involves intermolecular autophosphorylation (Glantschnig et al., 2002; Lee and Yonehara, 2002), and that activated Hpo phosphorylates Sav, Wts, and Mats (Wu et al., 2003; Wei et al., 2007). Sav binds to both Hpo and Wts, and is thought to act as a scaffolding protein that links them together (Wu et al., 2003). Mats is an essential co-factor for Wts (Lai et al., 2005). Wts activity is also regulated by Hpo-dependent phosphorylation, and Wts autophosphorylation (Wei et al., 2007).
Figure 1. Schematic model of the Hippo signaling network.
The model depicts most identified components of the Drosophila Hpo pathway, grouped into modules as discussed in the text. These include the Hpo kinase cassette (outlined in green), the Mer-ex complex (outlined in purple), the Fat branch of the pathway (outlined in orange), and transcription factors (within nucleus, shaded grey). Arrows depict key regulatory connections, pointed arrows show activating effects, block arrows show inhibiting effects.
Table 1.
Summary of Hippo pathway Components in Drosophila
| Drosophila Protein | Vertebrate homologues | Protein type or motifs | Role |
|---|---|---|---|
| Fat Pathway | |||
| Dachsous (Ds) | Dchs1, 2 | Atypical Cadherin | Ligand for Fat |
| Fat | Fat4 | Atypical Cadherin | Receptor |
| Lowfat (Lft) | Lix1, Lix1L | novel | Levels of Fat & Ds |
| Four-jointed (Fj) | Fjx1 | Golgi-localized kinase | Phosphorylates Fat & Ds cadherin domains |
| Discs overgrown (Dco) | CK1δ, ε | CK1 family Kinase | Phosphorylates Fat cytoplasmic domain |
| Dachs | ? | Unconventional Myosin | Transduces Signal from Fat to Wts |
| Approximated (App) | ZDHHC’s | Palmitoyltransferase | Dachs membrane localization |
| Zyx102 (Zyx) | Zyxin, Lpp, Trip6 | LIM domains | Interacts with Wts |
| Ex-Mer complex | |||
| Crumbs (Crb) | Crb1-3 | Transmembrane, EGF domains |
Receptor |
| Expanded (Ex) | FRMD6 (Willin) | FERM, PPXY domains | Regulates Hpo |
| Merlin (Mer) | Merlin | FERM domain | Regulates Hpo |
| Kibra (Kbr) | Kibra | WW, C2 domains | Regulates Hpo |
| Kinase Cassette | |||
| Hippo (Hpo) | Mst1,2 | Sterile-20 family Ser/Thr kinase |
Phosphorylates Wts |
| Salvador (Sav) | WW45 | WW, SARAH domains | Scaffolding protein |
| Warts (Wts) | Lats1,2 | NDR family Ser/Thr kinase |
Phosphorylates Yki |
| Mob as tumour suppressor (Mats) |
Mob1,2 | NDR kinase family co- factor |
Promotes Wts activity |
| Other regulators | |||
| RASSF | RASSF1-6 | Ras-association and SARAH domains |
Scaffolding protein? |
| PP2A (STRIPAK) | PP2A (STRIPAK) | phosphatase | Inhibits Hpo activity |
| Jub | Ajuba, Limd1, Wtip | LIM domains | Inhibits Wts activation |
| Lgl | Lgl | WD40 motif | Scaffolding protein? |
| Jnk | Jnk | Kinase | Activates Yki (indirectly?) |
| Myopic (Mop) | HD-PTP | Phosphatase, PPXY, Bro motifs |
Binds and inhibits Yki |
| Transcription Factors | |||
| Yorkie (Yki) | Yap, Taz | WW domains | Transcriptional co-activator |
| Wpb2 | Wpb2 | PPXY motifs | Transcriptional co-activator |
| Scalloped (Sd) | TEAD/TEF 1-4 | TEA-domain | DNA binding |
| Mad | Smad | MH domains | DNA binding |
| Homothorax (Hth) | Meis1-3 | Homeodomain | DNA binding |
| Teashirt (Tsh) | Tshz1-3 | Zn-finger | DNA binding |
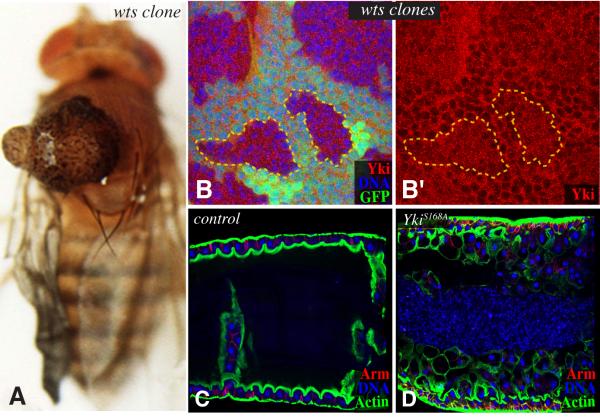
Figure 2. Examples of Hpo pathway phenotypes.
A) Adult fly with a wts mutant clone in the notum, resulting in a tumorous overgrowth (image courtesy of Tian Xu, see (Xu et al., 1995) for details). B) Portion of a wing imaginal disc with wts mutant clones, identified by lack of green staining; two examples are outlined by dashed lines. In wild-type cells, Yki (red) is predominantly cytoplasmic), but in wts mutant cells it is readily detected in the nucleus, resulting in uniform distribution of Yki (reproduced from Oh and Irvine, 2008). Panel B’ shows only the Yki stain. C) Close-up of the intestine of an adult fly. D) Close-up of the intestine of an adult fly in which an activated form of Yki (YkiS168A) is expressed within the differentiated cells. This stimulates an overproliferation of ISCs, and results in a massive overgrowth of the intestine (reproduced from Staley and Irvine, 2010).
Although multiple substrates for both Hpo and Wts kinases have been identified, the crucial target of Hpo is Wts, and the crucial target of Wts is a transcriptional co-activator protein, Yorkie (Yki) (Huang et al., 2005). Yki functions as an oncogene, as it promotes cell proliferation and growth, and inhibits cell death. Yki is phosphorylated by Wts on three Ser residues, amongst which the most critical is Ser168 (Dong et al., 2007; Oh and Irvine, 2008; Zhang et al., 2008; Oh and Irvine, 2009; Ren et al., 2010b). Phosphorylation of Ser168 creates a binding site for 14-3-3 proteins. This negatively regulates Yki activity by keeping it localized in the cytoplasm (Dong et al., 2007; Oh and Irvine, 2008; Zhang et al., 2008), potentially both through cytoplasmic anchoring and promoting nuclear export (Ren et al., 2010b). Fundamentally, one can think of the Hippo pathway as a constraint on Yki activity, which operates by preventing Yki accumulation within the nucleus (reviewed in Oh and Irvine, 2010) (Figure 2B).
Upstream regulators of the Hippo kinase cassette
A distinctive feature of Hippo signaling is the diverse and ever increasing number of upstream inputs that can feed into the core kinase cassette. Although our understanding of how they target the core kinase module remains incomplete, it’s clear that Hippo signaling functions as a network that integrates multiple upstream inputs, rather than a simple linear pathway (Fig. 1).
Mer-Ex complex
The first upstream regulators to be linked to the Hippo kinase cassette were two FERM (4.1, Ezrin, Radixin, Moesin) domain-containing proteins, Merlin (Mer) and Expanded (Ex) (Hamaratoglu et al., 2006). ex was initially identified as a tumor suppressor in Drosophila through mutations that cause overgrowth of the wing (Boedigheimer and Laughon, 1993). Mer, by contrast, was initially identified as a tumor suppressor in humans, through its association with Neurofibromatosis type 2 (NF2) (Gusella et al., 1996); subsequent studies showed that it also affects growth in Drosophila (LaJeunesse et al., 1998). Mer and ex are partially redundant, as mutation of either gene alone can result in increased Yki activation and increased tissue growth, but mutation of both genes together results in stronger phenotypes (McCartney et al., 2000; Hamaratoglu et al., 2006; Maitra et al., 2006). The respective contributions of Mer and ex to Hpo pathway regulation appear to vary, as mutation of ex alone results in stronger phenotypes than mutation of Mer alone in some organs, like the wing, but not in others, like the ovary (McCartney et al., 2000; Cho et al., 2006; Meignin et al., 2007; Polesello and Tapon, 2007; Yu et al., 2008; Zhao et al., 2008), and there are also temporal differences in their requirements between larval and pupal stages (Milton et al., 2010).
Although genetic studies indicate that Mer and Ex can each independently regulate Hippo signaling, they co-localize in epithelial cells, and can bind to each other (McCartney et al., 2000). More recently, Kibra was identified as a third partner in this Mer-Ex complex (Baumgartner et al., 2010; Genevet et al., 2010; Yu et al., 2010). kibra mutations cause overgrowth phenotypes comparable to those of Mer mutations, exhibit additive genetic interactions with Mer and ex, and Kibra protein co-localizes and physically associates with both Mer and Ex (Baumgartner et al., 2010; Genevet et al., 2010; Yu et al., 2010). Cell culture assays have revealed that Mer, Ex, and Kibra can promote phosphorylation of Hpo and Wts at sites associated with activation (Hamaratoglu et al., 2006; Yu et al., 2010). The biochemical details of how they effect Hpo and Wts activation have not been worked out, but multiple physical associations between members of the Ex-Mer-Kibra complex and the Hpo kinase cassette have been detected, including binding of Sav to Mer and Kibra, binding of Hpo to Ex, and binding of Wts to Kibra (Baumgartner et al., 2010; Genevet et al., 2010; Yu et al., 2010). In addition to influencing Hippo activity, Ex can also bind directly to Yki, and thereby tether Yki in the cytoplasm, providing a potential alternate mechanism for Yki regulation (Badouel et al., 2009; Oh et al., 2009).
FERM domain proteins are generally described as linking the F-actin cytoskeleton to cell membranes and membrane proteins (Mangeat et al., 1999), and recent discoveries emphasize the importance of these associations to Drosophila Mer and Ex function. Mutation of ex can increase the accumulation of apical F-actin, although this effect is not mediated through regulation of Yki (Fernandez et al., 2011; Sansores-Garcia et al., 2011). A phospho-lipid kinase, PI4KIIIalpha, was recently identified as a Drosophila gene with mutant phenotypes similar to those of Mer mutants (Yan et al., 2011). This requirement could be explained by the failure of Mer to localize to the apical membrane in mutant cells, presumably reflecting a requirement for PI4KIIIalpha in generating phospholipid binding sites for the Merlin FERM domain. This observation also emphasizes the importance of membrane localization to normal Mer function. Notably, localization of Kibra and Ex to the apical membrane was not disrupted in PI4KIIIalpha mutants. Kibra contains a C2 domain (Baumgartner et al., 2010; Genevet et al., 2010; Yu et al., 2010), a distinct phospho-lipid-binding motif that could potentially mediate its membrane association. Indeed, even though Mer, Ex, and Kibra can all associate with each other, each localizes to the membrane independently (McCartney et al., 2000; Baumgartner et al., 2010; Genevet et al., 2010; Yu et al., 2010).
Important insight into how Ex localization is regulated has come from the identification of Crumbs (Crb) as an Ex-binding protein and regulator of Hpo signaling (Chen et al., 2010; Grzeschik et al., 2010; Ling et al., 2010; Robinson et al., 2010). Crb is a transmembrane protein that localizes to the sub-apical membrane of epithelial cells, and in the Drosophila embryo functions together with other apical membrane complexes to organize apical-basal polarity (Tepass et al., 1990). Crb is not required for apical-basal polarity in Drosophila imaginal discs, but it was recently realized that loss of Crb is associated with an overgrowth phenotype (Chen et al., 2010; Ling et al., 2010; Robinson et al., 2010). Crb has a short intracellular domain that contains a juxtamembrane FERM-binding motif (FBM), and Crb can bind to Ex through its FBM. In crb mutant discs the sub-apical localization of Ex is greatly reduced, and Crb appears to regulate the Hippo pathway through this influence on Ex localization (Chen et al., 2010; Ling et al., 2010; Robinson et al., 2010). Notably, Crb may act as a transmembrane receptor recognizing cell-cell contacts through homophilic Crb-Crb binding, because localization of Crb and Ex to the membrane is lost even in wild-type cells, when they border crb mutant cells (Chen et al., 2010).
Prior to its linkage to Hippo signaling, Crb over-expression was reported to disrupt cell polarity and to promote cell proliferation in imaginal discs (Lu and Bilder, 2005). These two activities map to distinct regions of the Crb cytoplasmic domain: the polarity disruption is effected by a C-terminal PDZ-binding motif, while the proliferation phenotype maps to the FBM (Chen et al., 2010; Ling et al., 2010; Robinson et al., 2010). Crb over-expression is associated with Crb mis-localization, which drives mis-localization of Ex, and also appears to drive Ex turnover through an unknown mechanism (Chen et al., 2010; Grzeschik et al., 2010; Ling et al., 2010; Robinson et al., 2010). Thus, both too little and too much Crb results in Yki activation through inactivation of Ex.
Fat signaling
Fat is a large (more than five thousand amino acids) transmembrane protein with 34 cadherin repeats in its extracellular domain, and acts as a receptor for both Fat-Hippo signaling, and a distinct planar cell polarity pathway (reviewed in Reddy and Irvine, 2008). Null alleles of fat result in larval lethality, with overgrown imaginal discs (Mahoney et al., 1991). However, weak alleles of fat are viable and exhibit a broadening of the abdomen; this phenotype gives its name to fat. The tumor suppressor phenotype of Fat has been linked to Hippo signaling through several observations, including regulation of shared downstream target genes, genetic interactions, and influences on the levels and localization of Hippo pathway components, including Wts, Ex, and Yki (Bennett and Harvey, 2006; Cho et al., 2006; Silva et al., 2006; Willecke et al., 2006; Tyler and Baker, 2007; Oh and Irvine, 2008). Fat-Hpo signaling is genetically distinguishable from Hpo regulation by Ex/Mer/Kbr, and several genes have been identified as functioning specifically within the Fat branch of Hippo signaling (Fig. 1), including: Dachsous (Ds), an atypical cadherin that is a ligand for Fat (Cho and Irvine, 2004; Matakatsu and Blair, 2004; Cho et al., 2006; Matakatsu and Blair, 2006; Rogulja et al., 2008; Willecke et al., 2008; Brittle et al., 2010; Simon et al., 2010); Four-jointed (Fj), a Golgi-localized kinase that phosphorylates cadherin domains of Fat and Ds to modulate binding between them (Ishikawa et al., 2008; Brittle et al., 2010; Simon et al., 2010); Discs overgrown (Dco), a Drosophila casein kinase I that is involved in a Ds-dependent phosphorylation of the Fat cytoplasmic domain (Cho et al., 2006; Feng and Irvine, 2009; Sopko et al., 2009); Dachs, an unconventional myosin whose localization is regulated by Fat (Cho and Irvine, 2004; Cho et al., 2006; Mao et al., 2006); Approximated (App), a palmitoyltransferase required for Dachs membrane localization (Matakatsu and Blair, 2008); and Lowfat, a pioneer protein required for normal levels of Fat and Ds (Mao et al., 2009). Most recently, the LIM-domain protein Zyx102 (Zyx) was linked to Fat-Hippo signaling (Rauskolb et al., 2011). Fat signaling is crucial for Hippo pathway regulation in some tissues, such as imaginal discs and neuroepithelia, but dispensible in others such as the ovary (Bennett and Harvey, 2006; Cho et al., 2006; Silva et al., 2006; Willecke et al., 2006; Meignin et al., 2007; Polesello and Tapon, 2007; Reddy et al., 2010).
A unique feature of Fat signaling is its sensitivity to the slope and vector of the gradient of expression of the regulators Ds and Fj. Ds and Fj are normally expressed in gradients, and differences in their expression between neighboring cells stimulates Yki activity (Cho and Irvine, 2004; Cho et al., 2006; Rogulja et al., 2008; Willecke et al., 2008; Zecca and Struhl, 2010). These gradients are interpreted through effects on Dachs, whose membrane localization is regulated by Fj, Ds, and Fat (Cho et al., 2006; Mao et al., 2006; Rogulja et al., 2008; Willecke et al., 2008). Two mechanisms by which Dachs could influence Yki activity have been described. One involves a post-translational influence of Fat on Warts protein levels (Cho et al., 2006). Another involves an influence of Fat on the levels of Ex protein at the sub-apical membrane (Bennett and Harvey, 2006; Silva et al., 2006; Willecke et al., 2006; Feng and Irvine, 2007). Several observations, such as the rescue of fat mutants by Wts over-expression, the additive phenotypes of fat and ex mutants, and the lack of significant influence of dco or Zyx on Ex membrane localization (Feng and Irvine, 2007; Feng and Irvine, 2009; Rauskolb et al., 2011), suggest that the influence on Wts is the primary mechanism by which Fat regulates Hippo signaling. Nonetheless, the influence of Fat on Ex likely also contributes to Fat pathway phenotypes, and the respective contributions of these two mechanisms haven’t yet been clearly defined.
The Lethal giant larvae complex
Drosophila tumor suppressor mutations have been placed into two broad classes: hyperplastic tumor suppressors and neoplastic tumor suppressors, with neoplastic tumor suppressors distinguished by their disruption of apical-basal polarity and normal tissue architecture (Watson et al., 1994). Several of the classic hyperplastic tumor suppressors, including fat, ex, wts, and dco, were subsequently linked to Hippo signaling. Intriguingly, recent studies have now also established links between neoplastic tumor suppressors and Hippo signaling. Lethal giant larvae (Lgl) localizes to the basolateral membranes of epithelial cells, and acts in conjunction with two other proteins, Discs large (Dlg) and Scribble (Scrib) (Bilder et al., 2000; Humbert et al., 2008). This Lgl complex regulates apical-basal polarity by restricting the components of two other polarity complexes, the Crb complex (Crb, Stardust, and Patj) and the atypical Protein kinase C (aPKC) complex (aPKC, Bazooka, Par-6), to apical membranes.
Three recent studies, each examining distinct conditions, have identified and characterized links between the Lgl complex and the Hippo pathway (Grzeschik et al., 2010; Menendez et al., 2010; Sun and Irvine, 2011). Within the eye (but not the wing) imaginal disc, clones of cells mutant for lgl can retain apical-basal polarity, due to perdurance of the protein (Grzeschik et al., 2007). Nonetheless, these lgl mutant eye clones can overproliferate, and Grzeschik et al. (2010) observed that they are associated with Yki activation, and that Yki is required for the increased proliferation and suppression of apoptosis within lgl mutant clones. In polarity, Lgl acts antagonistically to aPKC, and the influence of Lgl on Yki activity could be suppressed by expression of dominant-negative aPKC. These authors also observed that both Hpo and a negative regulator of Hpo activity, Ras association family member (Rassf) (Polesello et al., 2006), are normally predominantly apical in eye disc cells, but become mis-localized laterally in cells mutant for lgl or expressing an activated form of aPKC. These observations led to the proposal that a novel Lgl-Hippo pathway affects Hpo signaling through an aPKC-mediated influence on Hpo and Rassf localization.
Sun and Irvine (2011) focused on an RNAi-mediated knock-down of lgl severe enough to disrupt apical-basal polarity in the wing disc. Consistent with Grzeschik et al. (2010), they found that the increased proliferation of lgl-depleted cells was associated with, and dependent upon, activation of Yki. However, Sun and Irvine (2011) identified an essential role for Jun kinase (Jnk) signaling in the activation of Yki within lgl-depleted cells. The mechanism by which Jnk signaling activates Yki is not yet known, but it can be suppressed by over-expression of Wts or Hpo. As discussed below, Jnk activation is also important for Yki regulation during regenerative growth (Karpowicz et al., 2010; Ren et al., 2010a; Shaw et al., 2010; Staley and Irvine, 2010; Sun and Irvine, 2011). Activation of aPKC could also activate Yki in wing discs, but this activation was also Jnk-dependent. Despite the common identification of Yki activation upon loss of lgl or activation of aPKC in these two studies, there are also differences. For example, Hpo appears to be broadly cytoplasmic in the wing disc (Genevet et al., 2009), rather than preferentially apical as in the eye disc (Grzeschik et al., 2010), and while direct activation of Jnk resulted in robust Yki activation in the wing disc, it had little effect in the eye disc (Sun and Irvine, 2011). Thus, there might be multiple mechanisms by which loss of Lgl results in Yki activation, and a better mechanistic understanding of how Lgl is linked to Yki regulation is needed.
When an entire imaginal disc is mutant for any member of the Lgl complex, a neoplastic tumor suppressor phenotype is observed – cells lose their apical basal polarity, and the discs overgrow. However, when small patches of mutant cells (clones) are made, the situation is more complex. These clones often die, but can survive, over-proliferate, and metastasize if an oncogenic mutation, such as activated-ras (rasv12) or Myc, is co-expressed (reviewed in Humbert et al., 2008). Menendez et al. (2010) investigated Yki activation in lgl mutant clones rescued by expression of rasv12. They described how the loss of lgl mutant clones could be ascribed to cell competition, and their rescue by rasv12 to a ras v12-promoted merging of clones, which effectively creates a microenvironment that suppresses cell competition. These authors also observed Yki activation specifically within the lgl rasv12 clones protected from cell competition. Common to all three of these studies is realization that a class of neoplastic tumor suppressors exemplified by lgl (dlg and scrib appear to act similarly) exert their influence on cell proliferation through an influence on the Hippo pathway.
F-actin
Another input into Yki regulation was identified recently with the discovery that mutation or down-regulation of either of the two subunits of Actin-capping protein (Cpa and Cpb) results in Yki activation (Fernandez et al., 2011; Sansores-Garcia et al., 2011). Capping proteins limit F-actin disassembly by binding the barbed ends of F-actin; consequently, Cpa or Cpb mutants have increased levels of F-actin (Janody and Treisman, 2006). The increased Yki activity associated with mutation or RNAi of Cpa or Cpb could be ascribed to increased F-actin, rather than a specific effect of Capping proteins, because increasing F-actin by mutation of capulet, or by expression of activated-Diaphanous, also increased Yki activity (Fernandez et al., 2011; Sansores-Garcia et al., 2011). Mutation of Cpa was previously shown to be associated with Jnk activation (Janody and Treisman, 2006), which raises the question of whether the influence of increased F-actin on Yki is mediated through Jnk. However, Sansores-Garcia et al (2011) observed that expression of a dominant negative isoform of Jnk (Basket, Bsk) only partially suppressed the overgrowth associated with RNAi of Cpa, and Yki activation was still detected. Thus, Jnk activation likely contributes to the influence of F-actin on Yki activation, but there may also be Jnk-independent effects of F-actin.
Diverse inputs for one pathway
A picture of Hpo signaling is now emerging in which diverse upstream inputs converge on the core kinase cassette, but regulate it in distinct ways (Fig. 1). One common theme is that key regulatory steps occur at the membrane. This is emphasized by the importance of membrane localization to the activity of upstream pathway components, such as Dachs, Mer, and Ex. Moreover, membrane localization of components of the core Hpo kinase cassette also appears to be important: in addition to the regulation of Hpo localization by Lgl in eye discs, and the identification of binding between core kinase cassette components and Mer, Ex and Kbr, studies of Mats revealed that a fraction of Mats normally localizes to the sub-apical membrane, and that forced membrane localization of Mats reduces tissue growth through increased Wts activity (Ho et al., 2010).
The responsiveness of Hpo signaling to distinct inputs begs the question of why these diverse upstream inputs should utilize a common response pathway. What they appear to share is a role in providing structural information about the cell and the local cellular milieu: apical-basal polarity, planar cell polarity, F-actin accumulation, and contact with neighboring cells. Indeed, even though there appear to be some differences between vertebrates and Drosophila in the specific identity of upstream Hpo pathway regulators, a common feature is the linkage of proteins associated with intercellular junctions and cell polarity.
Inhibitors of Hippo signaling
Most components of the Hippo pathway were first identified as tumor suppressor genes, and until recently yki and dachs were the only two genes in the pathway identified as mutations associated with reduced growth. This probably reflects the relative ease of identifying overgrowth mutants and assigning them to the pathway, as opposed to mutations associated with reduced growth, which is the expected phenotype for disruption of almost any essential cellular process. However, recently further progress has begun to be made on identifying additional components that act in opposition to the tumor suppressors of the Hpo pathway.
STRIPAK - PP2A complex
Given the central importance of protein kinases to Hpo signaling, it was to be expected that phosphatases should also play an important role, but until recently, none had been identified. Using both genomic and proteomic approaches, Ribeiro et al (2010) identified a PP2A phosphatase complex, STRIPAK (Striatin-interacting phosphatase and kinase), as both a Hpo-interacting complex, and a negative regulator of Hpo. This complex is related to mammalian STRIPAK complexes. Biochemical studies showed that depletion of the PP2A catalytic subunit Microtubule star (Mts) increased Hpo phosphorylation, implicating Hpo as a target of this phosphatase, and genetic studies of STRIPAK complex subunits were consistent with regulation of Hpo activity in vivo (Ribeiro et al., 2010). Intriguingly, the STRIPAK complex also associates with RASSF, a previously identified negative regulator of Hpo (Polesello et al., 2006), and binding experiments suggested that RASSF contributes to recruitment of STRIPAK to Hpo.
LIM domain proteins, Jub and Zyx
Recent studies have identified Ajuba LIM protein (Jub) and Zyx102 (Zyx) as novel components of Hippo signaling that act as negative regulators of Wts (Das Thakur et al., 2010; Rauskolb et al., 2011). These two proteins are structurally related, sharing three C-terminal LIM domains, but have distinct roles within the Hippo pathway. Jub is the sole Drosophila member of the Ajuba protein family, which in vertebrates includes Ajuba, LIMD1, and WTIP, whereas Zyx is the sole Drosophila member of the Zyxin family, which in vertebrates includes Zyxin, Lipoma Preferred Partner (LPP), and Thyroid-receptor interacting protein 6 (TRIP6). Ajuba family proteins are generally thought to link cell adhesive properties to nuclear responses (Marie et al., 2003; Langer et al., 2008), and can localize to adherens junctions. Zyxin family proteins have been implicated in cytoskeletal and transcriptional regulation (Wang and Gilmore, 2003; Grunewald et al., 2009), and Zyxin in particular has been implicated in linking mechanical stress to cell behavior (Hirata et al., 2008).
For both Ajuba and Zyxin family proteins, apparent redundancy among family members has been a challenge to defining their in vivo functions in vertebrates. Taking advantage of the existence of only a single family member in Drosophila, Das Thakur et al (2010) determined that mutation or RNAi of jub reduces growth of Drosophila tissues, and were able to link this phenotype to increased Hippo pathway activity. The exact mechanism by which the Jub proteins influence the Hpo kinase cascade is not clear, but genetic epistasis experiments positioned the action of Jub in between wts and hpo, and co-immunoprecipitation experiments identified interactions between Jub and both Sav and Wts. Experiments in cultured cells indicate that these interactions, and the ability of Jub to regulate Hpo signaling, are conserved in mammals (Das Thakur et al., 2010). In vertebrates, Jub proteins are recruited to adherens junctions upon cell-cell contact (Marie et al., 2003). Since Hpo signaling has been linked to contact inhibition in mammalian cells (Zhao et al., 2007), an intriguing possibility for the regulation of Jub is thus that its recruitment to junctions enhances Wts activity by preventing Jub from interacting with Wts and Sav. It’s also noteworthy that both Jub and Mer have been reported to bind to alpha-catenin in earlier studies (Marie et al., 2003; Gladden et al., 2010), as alpha-catenin was recently identified as a Hpo pathway regulator in mice (Schlegelmilch et al., 2011).
Zyx was identified as a novel component of the Hippo pathway through an RNAi-based genetic screen (Rauskolb et al., 2011). Like Jub, Zyx positively regulates Yki activity, and acts upstream of Wts. However, by contrast to Jub, which is required for both the Ex and Fat branches of Hippo signaling, Zyx appears to function specifically within the Fat branch, as Zyx RNAi effectively suppresses the fat tumor suppressor phenotype but not the ex tumor suppressor phenotype (Rauskolb et al., 2011). Moreover, a distinctive feature of the Fat branch of the Hippo pathway is its affects on Wts protein levels; further support for the placement of Zyx within the Fat branch of the pathway thus came from the observation that Zyx is required for the effect of fat on Wts levels. Binding and localization studies of Dachs and Zyx suggested a model for the role of Zyx in Fat-Hippo signal transduction. When Dachs and Zyx are at the membrane they co-localize. Fat activity antagonizes the localization of Dachs to the membrane (Mao et al., 2006), but it has not been clear how this influences downstream signaling. Rauskolb et al (2011) observed that Dachs stimulates Zyx-Wts binding, which suggests that the regulated localization of Dachs could serve to regulate Zyx-Wts binding. Whether Zyx homologues control Hippo signaling in vertebrates has not yet been determined. However, the ability of homologues of Zyx and Wts to associate with each other is conserved, and they can co-localize on the mitotic apparatus, where they influence mitosis (Hirota et al., 2000).
Additional Yki regulators
While much of the focus on Yki regulation has been devoted to processes that impinge on Wts-mediated phosphorylation of Yki, there has also been evidence that Yki can be regulated by direct binding to upstream components of the Hippo pathway, including Ex, Wts, and Hpo (Badouel et al., 2009; Oh et al., 2009). This binding is mediated through Yki’s WW domains, a protein-protein interaction domain that associates with short Proline-rich sequences, often within a PPXY motif (where X is any amino acid). In fact, interactions between WW domains and PPXY motifs are utilized repeatedly amongst components of the Hpo pathway, suggesting that competitive or cooperative interactions amongst proteins that share these motifs play an important role in Hpo pathway regulation (reviewed in Oh and Irvine, 2010). Recently, two additional PPXY-motif containing proteins, Wpb2 and Myopic (Mop) have been identified as proteins directly interacting with Yki through its WW domains, and as modulators of Hpo signaling (Gilbert et al., 2011; Zhang et al., 2011).
Mop is a Drosophila homolog of His-domain protein tyrosine phosphatase (HD-PTP), and was identified in a genetic screen for tumor suppressor mutations (Gilbert et al., 2011). On its own, mutation of mop results in cell death, but when apoptosis is blocked, mop mutations lead to Yki activation and overgrowth. When over-expressed, Mop can repress Yki activity, and it can do so by acting directly as a cytoplasmic anchor, rather than through modulating Yki phosphorylation. Mop is an endosomal protein, and the authors report that a substantial fraction of Yki is associated with endosomes, and that under mop mutant or RNAi conditions, Yki is shifted from late (rab7-positive) towards early (rab5-positive) endosomes. Another intriguing feature of mop mutations is that they are associated with upregulation of some downstream targets of Hpo signaling (eg ex), but not others (eg Diap1). The authors suggest that association of Yki with different endosomal compartments may influence interactions of Yki with binding partners, or post-translational modifications, which then influence Yki’s ability to regulate particular target genes. However, an alternative possibility is that mop mutations might downregulate Diap1 expression through an alternative process unrelated to Hpo signaling.
Wbp2 (WW domain binding protein-2) was first identified in mammals as a Yap-interacting protein (Chen et al., 1997), and recent studies have indicated that it can contribute to transcriptional activation by Yap and Taz (Dhananjayan et al., 2006; Chan et al., 2011), although the mechanism by which it does this is unknown. Studies of Drosophila wbp2 show that it acts as an enhancer of Yki’s transcriptional activity, but the phenotypes observed are quite mild compared to loss of yki (Zhang et al., 2011). Since the in vivo requirement for wbp2 has thus far only been addressed by RNAi, it’s possible that when mutations become available a more significant requirement will be revealed. The WW domains of Yki are required for Yki to stimulate transcription (Oh and Irvine, 2009; Zhang et al., 2009b), so if this requirement isn’t accounted for by Wbp2, there must be other, as yet unidentified, WW domain binding partners for Yki that enable it to activate transcription.
Another layer of Yki regulation was identified recently through the discovery that Myc, a key downstream transcriptional target of Yki, also effects a negative feedback regulation of Yki, through both transcriptional and post-transcriptional means (Neto-Silva et al., 2010). This Myc-dependent negative regulation of Yki could limit Myc-dependent overgrowths, whether induced by Yki activation or other growth control pathways that impinge on Myc regulation. The molecular details of how Myc regulates Yki levels are not yet known.
Transcriptional regulation by Yki
DNA-binding Yki partners
Yki is a transcriptional co-activator and lacks its own DNA binding domain. Instead, Yki functions in concert with DNA-binding transcription factors. The mammalian homologs of Yki (YAP and TAZ) had been reported to interact with several different DNA-binding proteins (reviewed in Bertini et al., 2009; Wang et al., 2009), but initially it wasn’t clear which, if any, of them might be linked to Hippo signaling. However, in 2008 three studies identified Drosophila Scalloped (Sd) as a DNA-binding partner for Yki in Hpo signaling, and identified Hpo-responsive enhancers within a known downstream target gene, Diap1 (Goulev et al., 2008; Wu et al., 2008; Zhang et al., 2008). A puzzling aspect of this discovery, however, is that yki appears to be required for the growth of all imaginal discs, whereas sd is principally required in the wing (Campbell et al., 1992; Liu et al., 2000). Since then, additional DNA-binding partners for Yki have been identified, including Mothers against Dpp (Mad) (Alarcon et al., 2009; Peng et al., 2009; Oh and Irvine, 2011) and a Homothrax (Hth) – Teashirt (Tsh) complex. Both Hth and Mad contribute to regulation of the growth promoting microRNA gene bantam (ban). Mad is best known as a transcription factor of the Dpp signaling pathway, and its identification as a Yki partner defines a crucial point of intersection between Hpo and Dpp signaling that is important for growth regulation in Drosophila (Oh and Irvine, 2011).
The existence of multiple DNA-binding partners for Yki provides for an unexpected diversity in the regulation of downstream transcriptional responses to Hpo signaling. It appears that in some cases this is deployed to provide tissue-specific responses to Hpo signaling. For example, Hth is a key Yki partner in the anterior eye disc (Peng et al., 2009), and Sd is a key Yki partner in the wing disc (Goulev et al., 2008; Wu et al., 2008; Zhang et al., 2008). In other cases it may expand the repertoire of downstream target genes regulated by Yki. For example, Diap1 is a target of a Yki-Sd transcription factor, but not a Yki-Mad or Yki-Hth transcription factor (Peng et al., 2009; Oh and Irvine, 2011). A recent study emphasizes that downstream transcriptional programs can also be modified through cooperative interactions between Yki complexes and other transcription factor complexes (Nicolay et al., 2011). These authors found that the ability of Yki to bypass cell cycle exit, and thereby promote continuous cell proliferation, was dependent upon co-regulation of downstream genes by Yki-Sd together with E2F1.
Downstream targets of Hippo signaling
Several downstream targets of Yki that are important for its role in growth control have been identified, including promoters of growth like ban and Myc (Nolo et al., 2006; Thompson and Cohen, 2006; Neto-Silva et al., 2010; Ziosi et al., 2010), promoters of cell cycle progression like E2F1 and cyclins A,B and E (Tapon et al., 2002; Silva et al., 2006; Tyler and Baker, 2007; Goulev et al., 2008), and inhibitors of apoptosis like Diap1 (Wu et al., 2003). Although not all of these are known to be direct targets, there is evidence that at least Myc, Diap1 and ban are directly regulated by Yki, through identification of binding sites for DNA-binding partners of Yki (Wu et al., 2008; Zhang et al., 2008; Peng et al., 2009; Neto-Silva et al., 2010; Oh and Irvine, 2011). Another important class of targets are components of other signaling pathways, including ligands for the Notch, Wnt, EGFR and Jak-Stat pathways (Cho et al., 2006; Zhang et al., 2009a; Karpowicz et al., 2010; Ren et al., 2010a; Shaw et al., 2010; Staley and Irvine, 2010), as this provides an important point of cross talk between pathways. Additionally, the glypicans Dally and Dally-like, which influence the extracellular levels and signaling efficiency of ligands for the Hedgehog, BMP and Wnt pathways, are also regulated by Hpo signaling (Baena-Lopez et al., 2008). Finally, several components of the Hpo pathway, including Ex, Mer, Kibra, Crb, and Fj, are also downstream targets (Cho et al., 2006; Hamaratoglu et al., 2006; Genevet et al., 2009; Genevet et al., 2010). Their regulation provides for a negative feedback regulation, which is an important feature of many signaling pathways.
The Hippo Pathway and Regeneration
While the Hpo pathway has become well known for its contributions to growth control during normal development, recent studies have emphasized that the Hippo pathway also plays an important role during regenerative growth. The ability of some animals to regenerate damaged or removed body parts has long intrigued scientists, and recently the Hippo pathway has been investigated in two different Drosophila regeneration models (discussed below), as well as a Cricket model (Bando et al., 2009). Undifferentiated Drosophila imaginal discs can replace damaged or missing parts through increased proliferation to replace the missing tissue. Indeed an ability of dying cells to stimulate proliferation of their neighbors, referred to as compensatory cell proliferation, was first identified and characterized within imaginal discs (reviewed in Fan and Bergmann, 2008). Two recent studies showed that Yki is hyperactivated within Drosophila wing discs in response to tissue damage, whether induced surgically, genetically, or by irradiation (Grusche et al., 2011; Sun and Irvine, 2011). This Yki activation appears to be important for compensatory cell proliferation, and intriguingly, regenerating tissues exhibit a high requirement for Yki activity, because animals with only one wild-type copy of Yki will develop normally, whereas their ability to effect disc regeneration is substantially impaired (Grusche et al., 2011; Sun and Irvine, 2011). Mechanisms that can contribute to Yki regulation during normal development, such as the Fat-Hippo pathway, can also contribute to Yki regulation during regenerative growth, but are not sufficient to account for Yki hyperactivation (Grusche et al., 2011). Instead, the Jnk pathway, which has been studied in Drosophila for its roles in damage response, regeneration, and compensatory cell proliferation (reviewed in Fan and Bergmann, 2008; Bergantinos et al., 2010), is crucial for Yki activation in response to tissue damage (Sun and Irvine, 2011). Because the Drosophila Jnk, Bsk, is required for Yki activation during regeneration, but not during normal development (Igaki, 2009), Jnk signaling is a context-specific input into the Hpo pathway.
Within the past several years, the adult midgut of Drosophila has been established as a model organ for studying stem cell-based adult tissue homeostasis (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006). Just as in vertebrates, the entire midgut normally turns over continually, through a basal rate of intestinal stem cell (ISC) proliferation and differentiation of non-stem cell daughters. However, when fly intestines are exposed to toxins or pathogens, probably a common occurrence in nature given their diet of microbe-containing rotting fruits, then the differentiated cells of the intestine can be damaged, and proliferation of ISCs increases to replace them. As a regeneration model, it exhibits some distinguishing features from imaginal discs, including a true regeneration of adult tissues, as opposed to replacement of undifferentiated cells within a developing organ, and a stem-cell based proliferation mechanism. Over the past few years several groups had described how tissue damage and infection is conveyed largely through Jnk signaling, and ISC proliferation and differentiation effected through Jak-Stat signaling (Amcheslavsky et al., 2009; Beebe et al., 2009; Buchon et al., 2009; Chatterjee and Ip, 2009; Jiang et al., 2009). Now, the Hippo pathway has been linked to these regulatory mechanisms, and shown to play a central role in controlling intestinal homeostasis (Karpowicz et al., 2010; Ren et al., 2010a; Shaw et al., 2010; Staley and Irvine, 2010).
These studies showed that damage to the intestine, mediated by toxins or bacterial infection, results in Yki activation, and Yki activation is required for elevated ISC proliferation in response to damage (Karpowicz et al., 2010; Ren et al., 2010a; Shaw et al., 2010; Staley and Irvine, 2010). Moreover, Yki provides a crucial link between the damage-sensing Jnk pathway, and the Jak-Stat pathway, which promotes stem cell proliferation. Jnk can directly activate Yki in differentiated intestinal cells, and Yki activation in these cells induces expression of Jak-Stat pathway ligands (Upd proteins), and also the EGFR ligand Vein. These observations define a crucial non-autonomous role for Yki in promoting cell proliferation in the intestine. Three groups also described an autonomous role for Yki within the ISCs in promoting their proliferation (Karpowicz et al., 2010; Ren et al., 2010a; Shaw et al., 2010), which was not observed in our experiments (Staley and Irvine, 2010). A complicating factor is that the ISCs are the progenitors of the differentiated cells, and thus even when the promoters used to drive expression of transgenes are specific for ISCs, the resulting gene products or small RNAs could perdure into differentiated cells. Thus, in our view additional experiments would be required to clearly establish that Yki acts within the ISCs themselves to promote proliferation, in addition to its role in the differentiated cells. Interestingly, Yap is also required for mouse intestine regeneration after exposure to damaging toxins (Cai et al., 2010).
Future Directions
The pace of discovery in the Hpo signaling field over the past few years has been remarkable, and because of its many advantages for gene discovery and characterization, research in Drosophila has continued to play a leading role. Substantial progress in identifying pathway components has been made recently, but we still lack a mechanistic understanding of crucial steps in the pathway, such as how the activity of the Hpo kinase is controlled by upstream regulators, or how the levels of Wts protein are controlled by Dachs and Zyx. Attaining these mechanistic insights will require substantial investments in both biochemical approaches (eg to reconstitute key steps in signal transduction in vitro), and better reagents for imaging steps in signal transduction in vivo. Reagents that would enable discrete identification and localization of active versus inactive isoforms of pathway components would be especially valuable. In short, in a field that has been dominated by geneticists, opportunities for talented biochemists and cell biologists abound.
One of the remarkable features of the pathway is the number of different inputs into upstream regulation. A key challenge for the future is to understand how all of the different inputs into the pathway are related to each other. A further challenge will be to understand how diverse inputs are integrated to achieve appropriate levels of Yki activity. Another continuing challenge is to understand how the Hpo pathway is integrated with other growth control pathways. Some progress has been made, for example with the discoveries of the regulation of Yki by Myc, the regulation of ligands for other pathways by Yki, and the co-regulation of ban by Yki and Mad, but important work remains to be done. For example, Wnt signaling also plays important roles in tissue regeneration, and there is evidence for cross talk between Hpo and Wnt signaling (Varelas et al., 2010), but we don’t yet have a clear understanding of how these pathways are integrated to regulate organ growth and regeneration.
Another striking feature of Hpo signaling is the potential for great diversity in transcriptional responses, through the ability of Yki to associate with different DNA-binding partners. To date most research has focused on a limited set of partners and downstream target genes, and the full extent and cell type diversity of transcriptional responses to Yki activity remains an open question.
ACKNOWLEDGEMENTS
We thank C. Rauskolb for comments of the manuscript, and Tian Xu for providing the image of a fly with a wts clone. Research in the Irvine lab is supported by the Howard Hughes Medical Institute, NIH grant 2R01 GM078620, and HFSP grant RGP0016/2010.
References
- Alarcon C, Zaromytidou AI, Xi Q, Gao S, Yu J, Fujisawa S, Barlas A, Miller AN, Manova-Todorova K, Macias MJ, Sapkota G, Pan D, Massague J. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell. 2009;139:757–769. doi: 10.1016/j.cell.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amcheslavsky A, Jiang J, Ip YT. Tissue damage-induced intestinal stem cell division in Drosophila. Cell Stem Cell. 2009;4:49–61. doi: 10.1016/j.stem.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badouel C, Gardano L, Amin N, Garg A, Rosenfeld R, Le Bihan T, McNeill H. The FERM-domain protein Expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Dev Cell. 2009;16:411–420. doi: 10.1016/j.devcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- Baena-Lopez LA, Rodriguez I, Baonza A. The tumor suppressor genes dachsous and fat modulate different signalling pathways by regulating dally and dally-like. Proc Natl Acad Sci U S A. 2008;105:9645–9650. doi: 10.1073/pnas.0803747105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bando T, Mito T, Maeda Y, Nakamura T, Ito F, Watanabe T, Ohuchi H, Noji S. Regulation of leg size and shape by the Dachsous/Fat signalling pathway during regeneration. Development. 2009;136:2235–2245. doi: 10.1242/dev.035204. [DOI] [PubMed] [Google Scholar]
- Baumgartner R, Poernbacher I, Buser N, Hafen E, Stocker H. The WW domain protein Kibra acts upstream of Hippo in Drosophila. Dev Cell. 2010;18:309–316. doi: 10.1016/j.devcel.2009.12.013. [DOI] [PubMed] [Google Scholar]
- Beebe K, Lee W, Micchelli CA. JAK/STAT signaling coordinates stem cell proliferation and multilineage differentiation in the Drosophila intestinal stem cell lineage. Dev Biol. 2009 doi: 10.1016/j.ydbio.2009.10.045. [DOI] [PubMed] [Google Scholar]
- Bennett FC, Harvey KF. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr Biol. 2006;16:2101–2110. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- Bergantinos C, Vilana X, Corominas M, Serras F. Imaginal discs: Renaissance of a model for regenerative biology. Bioessays. 2010;32:207–217. doi: 10.1002/bies.200900105. [DOI] [PubMed] [Google Scholar]
- Bertini E, Oka T, Sudol M, Strano S, Blandino G. YAP: at the crossroad between transformation and tumor suppression. Cell Cycle. 2009;8:49–57. doi: 10.4161/cc.8.1.7259. [DOI] [PubMed] [Google Scholar]
- Bilder D, Li M, Perrimon N. Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science. 2000;289:113–116. doi: 10.1126/science.289.5476.113. [DOI] [PubMed] [Google Scholar]
- Boedigheimer M, Laughon A. Expanded: a gene involved in the control of cell proliferation in imaginal discs. Development. 1993;118:1291–1301. doi: 10.1242/dev.118.4.1291. [DOI] [PubMed] [Google Scholar]
- Brittle AL, Repiso A, Casal J, Lawrence PA, Strutt D. Four-jointed modulates growth and planar polarity by reducing the affinity of dachsous for fat. Curr Biol. 2010;20:803–810. doi: 10.1016/j.cub.2010.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 2009;23:2333–2344. doi: 10.1101/gad.1827009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Zhang N, Zheng Y, de Wilde RF, Maitra A, Pan D. The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev. 2010;24:2383–2388. doi: 10.1101/gad.1978810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S, Inamdar M, Rodrigues V, Raghavan V, Palazzolo M, Chovnick A. The scalloped gene encodes a novel, evolutionarily conserved transcription factor required for sensory organ differentiation in Drosophila. Genes Dev. 1992;6:367–379. doi: 10.1101/gad.6.3.367. [DOI] [PubMed] [Google Scholar]
- Chan SW, Lim CJ, Huang C, Chong YF, Gunaratne HJ, Hogue KA, Blackstock WP, Harvey KF, Hong W. WW domain-mediated interaction with Wbp2 is important for the oncogenic property of TAZ. Oncogene. 2011;30:600–610. doi: 10.1038/onc.2010.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee M, Ip YT. Pathogenic stimulation of intestinal stem cell response in Drosophila. J Cell Physiol. 2009;220:664–671. doi: 10.1002/jcp.21808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Gajewski KM, Hamaratoglu F, Bossuyt W, Sansores-Garcia L, Tao C, Halder G. The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc Natl Acad Sci U S A. 2010;107:15810–15815. doi: 10.1073/pnas.1004060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HI, Einbond A, Kwak SJ, Linn H, Koepf E, Peterson S, Kelly JW, Sudol M. Characterization of the WW domain of human yes-associated protein and its polyproline-containing ligands. The Journal of biological chemistry. 1997;272:17070–17077. doi: 10.1074/jbc.272.27.17070. [DOI] [PubMed] [Google Scholar]
- Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a Fat tumor suppressor pathway. Nat Genet. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- Cho E, Irvine KD. Action of fat, four-jointed, dachsous and dachs in distal-to-proximal wing signaling. Development. 2004;131:4489–4500. doi: 10.1242/dev.01315. [DOI] [PubMed] [Google Scholar]
- Das Thakur M, Feng Y, Jagannathan R, Seppa MJ, Skeath JB, Longmore GD. Ajuba LIM proteins are negative regulators of the Hippo signaling pathway. Curr Biol. 2010;20:657–662. doi: 10.1016/j.cub.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhananjayan SC, Ramamoorthy S, Khan OY, Ismail A, Sun J, Slingerland J, O’Malley BW, Nawaz Z. WW domain binding protein-2, an E6-associated protein interacting protein, acts as a coactivator of estrogen and progesterone receptors. Molecular endocrinology. 2006;20:2343–2354. doi: 10.1210/me.2005-0533. [DOI] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Bergmann A. Apoptosis-induced compensatory proliferation. The Cell is dead. Long live the Cell! Trends Cell Biol. 2008;18:467–473. doi: 10.1016/j.tcb.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Irvine KD. Fat and expanded act in parallel to regulate growth through warts. Proc Natl Acad Sci U S A. 2007;104:20362–20367. doi: 10.1073/pnas.0706722105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Irvine KD. Processing and phosphorylation of the Fat receptor. Proc Natl Acad Sci U S A. 2009;106:11989–11994. doi: 10.1073/pnas.0811540106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez BG, Gaspar P, Bras-Pereira C, Jezowska B, Rebelo SR, Janody F. Actin-Capping Protein and the Hippo pathway regulate F-actin and tissue growth in Drosophila. Development. 2011;138:2337–2346. doi: 10.1242/dev.063545. [DOI] [PubMed] [Google Scholar]
- Genevet A, Polesello C, Blight K, Robertson F, Collinson LM, Pichaud F, Tapon N. The Hippo pathway regulates apical-domain size independently of its growth-control function. J Cell Sci. 2009;122:2360–2370. doi: 10.1242/jcs.041806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genevet A, Wehr MC, Brain R, Thompson BJ, Tapon N. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev Cell. 2010;18:300–308. doi: 10.1016/j.devcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MM, Tipping M, Veraksa A, Moberg KH. A Screen for Conditional Growth Suppressor Genes Identifies the Drosophila Homolog of HD-PTP as a Regulator of the Oncoprotein Yorkie. Developmental Cell. 2011;20:700–712. doi: 10.1016/j.devcel.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladden AB, Hebert AM, Schneeberger EE, McClatchey AI. The NF2 tumor suppressor, Merlin, regulates epidermal development through the establishment of a junctional polarity complex. Developmental Cell. 2010;19:727–739. doi: 10.1016/j.devcel.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glantschnig H, Rodan GA, Reszka AA. Mapping of MST1 kinase sites of phosphorylation. Activation and autophosphorylation. J Biol Chem. 2002;277:42987–42996. doi: 10.1074/jbc.M208538200. [DOI] [PubMed] [Google Scholar]
- Goulev Y, Fauny JD, Gonzalez-Marti B, Flagiello D, Silber J, Zider A. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr Biol. 2008;18:435–441. doi: 10.1016/j.cub.2008.02.034. [DOI] [PubMed] [Google Scholar]
- Grunewald TG, Pasedag SM, Butt E. Cell Adhesion and Transcriptional Activity - Defining the Role of the Novel Protooncogene LPP. Transl Oncol. 2009;2:107–116. doi: 10.1593/tlo.09112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grusche FA, Degoutin JL, Richardson HE, Harvey KF. The Salvador/Warts/Hippo pathway controls regenerative tissue growth in Drosophila melanogaster. Developmental biology. 2011;350:255–266. doi: 10.1016/j.ydbio.2010.11.020. [DOI] [PubMed] [Google Scholar]
- Grzeschik NA, Amin N, Secombe J, Brumby AM, Richardson HE. Abnormalities in cell proliferation and apico-basal cell polarity are separable in Drosophila lgl mutant clones in the developing eye. Developmental biology. 2007;311:106–123. doi: 10.1016/j.ydbio.2007.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzeschik NA, Parsons LM, Allott ML, Harvey KF, Richardson HE. Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr Biol. 2010;20:573–581. doi: 10.1016/j.cub.2010.01.055. [DOI] [PubMed] [Google Scholar]
- Gusella JF, Ramesh V, MacCollin M, Jacoby LB. Neurofibromatosis 2: loss of merlin’s protective spell. Curr Opin Genet Dev. 1996;6:87–92. doi: 10.1016/s0959-437x(96)90016-7. [DOI] [PubMed] [Google Scholar]
- Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, Halder G. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- Hirata H, Tatsumi H, Sokabe M. Zyxin emerges as a key player in the mechanotransduction at cell adhesive structures. Commun Integr Biol. 2008;1:192–195. doi: 10.4161/cib.1.2.7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Morisaki T, Nishiyama Y, Marumoto T, Tada K, Hara T, Masuko N, Inagaki M, Hatakeyama K, Saya H. Zyxin, a regulator of actin filament assembly, targets the mitotic apparatus by interacting with h-warts/LATS1 tumor suppressor. J Cell Biol. 2000;149:1073–1086. doi: 10.1083/jcb.149.5.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho LL, Wei X, Shimizu T, Lai ZC. Mob as tumor suppressor is activated at the cell membrane to control tissue growth and organ size in Drosophila. Dev Biol. 2010;337:274–283. doi: 10.1016/j.ydbio.2009.10.042. [DOI] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Humbert PO, Grzeschik NA, Brumby AM, Galea R, Elsum I, Richardson HE. Control of tumourigenesis by the Scribble/Dlg/Lgl polarity module. Oncogene. 2008;27:6888–6907. doi: 10.1038/onc.2008.341. [DOI] [PubMed] [Google Scholar]
- Igaki T. Correcting developmental errors by apoptosis: lessons from Drosophila JNK signaling. Apoptosis : an international journal on programmed cell death. 2009;14:1021–1028. doi: 10.1007/s10495-009-0361-7. [DOI] [PubMed] [Google Scholar]
- Ishikawa HO, Takeuchi H, Haltiwanger RS, Irvine KD. Four-jointed is a Golgi kinase that phosphorylates a subset of cadherin domains. Science. 2008;321:401–404. doi: 10.1126/science.1158159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janody F, Treisman JE. Actin capping protein alpha maintains vestigial-expressing cells within the Drosophila wing disc epithelium. Development. 2006;133:3349–3357. doi: 10.1242/dev.02511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Zhang W, Wang B, Trinko R, Jiang J. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 2003;17:2514–2519. doi: 10.1101/gad.1134003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137:1343–1355. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- Kango-Singh M, Nolo R, Tao C, Verstreken P, Hiesinger PR, Bellen HJ, Halder G. Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development. 2002;129:5719–5730. doi: 10.1242/dev.00168. [DOI] [PubMed] [Google Scholar]
- Kango-Singh M, Singh A. Regulation of organ size: insights from the Drosophila Hippo signaling pathway. Dev Dyn. 2009;238:1627–1637. doi: 10.1002/dvdy.21996. [DOI] [PubMed] [Google Scholar]
- Karpowicz P, Perez J, Perrimon N. The Hippo tumor suppressor pathway regulates intestinal stem cell regeneration. Development. 2010;137:4135–4145. doi: 10.1242/dev.060483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai ZC, Wei X, Shimizu T, Ramos E, Rohrbaugh M, Nikolaidis N, Ho LL, Li Y. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- LaJeunesse DR, McCartney BM, Fehon RG. Structural analysis of Drosophila merlin reveals functional domains important for growth control and subcellular localization. J Cell Biol. 1998;141:1589–1599. doi: 10.1083/jcb.141.7.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer EM, Feng Y, Zhaoyuan H, Rauscher FJ, 3rd, Kroll KL, Longmore GD. Ajuba LIM proteins are snail/slug corepressors required for neural crest development in Xenopus. Dev Cell. 2008;14:424–436. doi: 10.1016/j.devcel.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Yonehara S. Phosphorylation and dimerization regulate nucleocytoplasmic shuttling of mammalian STE20-like kinase (MST) J Biol Chem. 2002;277:12351–12358. doi: 10.1074/jbc.M108138200. [DOI] [PubMed] [Google Scholar]
- Ling C, Zheng Y, Yin F, Yu J, Huang J, Hong Y, Wu S, Pan D. The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc Natl Acad Sci U S A. 2010;107:10532–10537. doi: 10.1073/pnas.1004279107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Grammont M, Irvine KD. Roles for scalloped and vestigial in regulating cell affinity and interactions between the wing blade and the wing hinge. Dev Biol. 2000;228:287–303. doi: 10.1006/dbio.2000.9939. [DOI] [PubMed] [Google Scholar]
- Lu H, Bilder D. Endocytic control of epithelial polarity and proliferation in Drosophila. Nature cell biology. 2005;7:1232–1239. doi: 10.1038/ncb1324. [DOI] [PubMed] [Google Scholar]
- Mahoney PA, Weber U, Onofrechuk P, Biessmann H, Bryant PJ, Goodman CS. The fat tumor suppressor gene in Drosophila encodes a novel member of the cadherin gene superfamily. Cell. 1991;67:853–868. doi: 10.1016/0092-8674(91)90359-7. [DOI] [PubMed] [Google Scholar]
- Maitra S, Kulikauskas RM, Gavilan H, Fehon RG. The tumor suppressors Merlin and expanded function cooperatively to modulate receptor endocytosis and signaling. Curr Biol. 2006;16:702–709. doi: 10.1016/j.cub.2006.02.063. [DOI] [PubMed] [Google Scholar]
- Mangeat P, Roy C, Martin M. ERM proteins in cell adhesion and membrane dynamics. Trends Cell Biol. 1999;9:187–192. doi: 10.1016/s0962-8924(99)01544-5. [DOI] [PubMed] [Google Scholar]
- Mao Y, Kucuk B, Irvine KD. Drosophila lowfat, a novel modulator of Fat signaling. Development. 2009;136:3223–3233. doi: 10.1242/dev.036152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Rauskolb C, Cho E, Hu WL, Hayter H, Minihan G, Katz FN, Irvine KD. Dachs: an unconventional myosin that functions downstream of Fat to regulate growth, affinity and gene expression in Drosophila. Development. 2006;133:2539–2551. doi: 10.1242/dev.02427. [DOI] [PubMed] [Google Scholar]
- Marie H, Pratt SJ, Betson M, Epple H, Kittler JT, Meek L, Moss SJ, Troyanovsky S, Attwell D, Longmore GD, Braga VM. The LIM protein Ajuba is recruited to cadherin-dependent cell junctions through an association with alpha-catenin. J Biol Chem. 2003;278:1220–1228. doi: 10.1074/jbc.M205391200. [DOI] [PubMed] [Google Scholar]
- Matakatsu H, Blair SS. Interactions between Fat and Dachsous and the regulation of planar cell polarity in the Drosophila wing. Development. 2004;131:3785–3794. doi: 10.1242/dev.01254. [DOI] [PubMed] [Google Scholar]
- Matakatsu H, Blair SS. Separating the adhesive and signaling functions of the Fat and Dachsous protocadherins. Development. 2006;133:2315–2324. doi: 10.1242/dev.02401. [DOI] [PubMed] [Google Scholar]
- Matakatsu H, Blair SS. The DHHC palmitoyltransferase approximated regulates Fat signaling and Dachs localization and activity. Curr Biol. 2008;18:1390–1395. doi: 10.1016/j.cub.2008.07.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney BM, Kulikauskas RM, LaJeunesse DR, Fehon RG. The neurofibromatosis-2 homologue, Merlin, and the tumor suppressor expanded function together in Drosophila to regulate cell proliferation and differentiation. Development. 2000;127:1315–1324. doi: 10.1242/dev.127.6.1315. [DOI] [PubMed] [Google Scholar]
- Meignin C, Alvarez-Garcia I, Davis I, Palacios IM. The salvador-warts-hippo pathway is required for epithelial proliferation and axis specification in Drosophila. Curr Biol. 2007;17:1871–1878. doi: 10.1016/j.cub.2007.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez J, Perez-Garijo A, Calleja M, Morata G. A tumor-suppressing mechanism in Drosophila involving cell competition and the Hippo pathway. Proc Natl Acad Sci U S A. 2010;107:14651–14656. doi: 10.1073/pnas.1009376107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micchelli CA, Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–479. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- Milton CC, Zhang X, Albanese NO, Harvey KF. Differential requirement of Salvador-Warts-Hippo pathway members for organ size control in Drosophila melanogaster. Development. 2010;137:735–743. doi: 10.1242/dev.042309. [DOI] [PubMed] [Google Scholar]
- Neto-Silva RM, de Beco S, Johnston LA. Evidence for a growth-stabilizing regulatory feedback mechanism between Myc and Yorkie, the Drosophila homolog of Yap. Dev Cell. 2010;19:507–520. doi: 10.1016/j.devcel.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolay BN, Bayarmagnai B, Islam AB, Lopez-Bigas N, Frolov MV. Cooperation between dE2F1 and Yki/Sd defines a distinct transcriptional program necessary to bypass cell cycle exit. Genes & development. 2011;25:323–335. doi: 10.1101/gad.1999211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolo R, Morrison CM, Tao C, Zhang X, Halder G. The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr Biol. 2006;16:1895–1904. doi: 10.1016/j.cub.2006.08.057. [DOI] [PubMed] [Google Scholar]
- Oh H, Irvine KD. In vivo regulation of Yorkie phosphorylation and localization. Development. 2008;135:1081–1088. doi: 10.1242/dev.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Irvine KD. In vivo analysis of Yorkie phosphorylation sites. Oncogene. 2009;28:1916–1927. doi: 10.1038/onc.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Irvine KD. Yorkie: the final destination of Hippo signaling. Trends Cell Biol. 2010;20:410–417. doi: 10.1016/j.tcb.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Irvine KD. Cooperative regulation of growth by Yorkie and Mad through bantam. Dev Cell. 2011;20:109–122. doi: 10.1016/j.devcel.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Reddy BV, Irvine KD. Phosphorylation-independent repression of Yorkie in Fat-Hippo signaling. Dev Biol. 2009;335:188–197. doi: 10.1016/j.ydbio.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature. 2006;439:470–474. doi: 10.1038/nature04333. [DOI] [PubMed] [Google Scholar]
- Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantalacci S, Tapon N, Leopold P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat Cell Biol. 2003;5:921–927. doi: 10.1038/ncb1051. [DOI] [PubMed] [Google Scholar]
- Peng HW, Slattery M, Mann RS. Transcription factor choice in the Hippo signaling pathway: homothorax and yorkie regulation of the microRNA bantam in the progenitor domain of the Drosophila eye imaginal disc. Genes Dev. 2009;23:2307–2319. doi: 10.1101/gad.1820009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesello C, Huelsmann S, Brown NH, Tapon N. The Drosophila RASSF homolog antagonizes the hippo pathway. Curr Biol. 2006;16:2459–2465. doi: 10.1016/j.cub.2006.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesello C, Tapon N. Salvador-warts-hippo signaling promotes Drosophila posterior follicle cell maturation downstream of notch. Curr Biol. 2007;17:1864–1870. doi: 10.1016/j.cub.2007.09.049. [DOI] [PubMed] [Google Scholar]
- Rauskolb C, Pan G, Reddy BV, Oh H, Irvine KD. Zyxin links fat signaling to the hippo pathway. PLoS biology. 2011;9:e1000624. doi: 10.1371/journal.pbio.1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy BV, Irvine KD. The Fat and Warts signaling pathways: new insights into their regulation, mechanism and conservation. Development. 2008;135:2827–2838. doi: 10.1242/dev.020974. [DOI] [PubMed] [Google Scholar]
- Reddy BVVG, Rauskolb C, Irvine KD. Influence of Fat-Hippo and Notch signaling on the proliferation and differentiation of Drosophila optic neuroepithelia. Development. 2010;137:2397–2408. doi: 10.1242/dev.050013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren F, Wang B, Yue T, Yun EY, Ip YT, Jiang J. Hippo signaling regulates Drosophila intestine stem cell proliferation through multiple pathways. Proc Natl Acad Sci U S A. 2010a;107:21064–21069. doi: 10.1073/pnas.1012759107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren F, Zhang L, Jiang J. Hippo signaling regulates Yorkie nuclear localization and activity through 14-3-3 dependent and independent mechanisms. Dev Biol. 2010b;337:303–312. doi: 10.1016/j.ydbio.2009.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro PS, Josue F, Wepf A, Wehr MC, Rinner O, Kelly G, Tapon N, Gstaiger M. Combined functional genomic and proteomic approaches identify a PP2A complex as a negative regulator of Hippo signaling. Mol Cell. 2010;39:521–534. doi: 10.1016/j.molcel.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Robinson BS, Huang J, Hong Y, Moberg KH. Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein Expanded. Curr Biol. 2010;20:582–590. doi: 10.1016/j.cub.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogulja D, Rauskolb C, Irvine KD. Morphogen control of wing growth through the Fat signaling pathway. Dev Cell. 2008;15:309–321. doi: 10.1016/j.devcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansores-Garcia L, Bossuyt W, Wada K, Yonemura S, Tao C, Sasaki H, Halder G. Modulating F-actin organization induces organ growth by affecting the Hippo pathway. The EMBO journal. 2011;30:2325–2335. doi: 10.1038/emboj.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D, Kreger BT, Vasioukhin V, Avruch J, Brummelkamp TR, Camargo FD. Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell. 2011;144:782–795. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RL, Kohlmaier A, Polesello C, Veelken C, Edgar BA, Tapon N. The Hippo pathway regulates intestinal stem cell proliferation during Drosophila adult midgut regeneration. Development. 2010;137:4147–4158. doi: 10.1242/dev.052506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr Biol. 2006;16:2081–2089. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Simon MA, Xu A, Ishikawa HO, Irvine KD. Modulation of Fat:Dachsous binding by the cadherin domain kinase four-jointed. Curr Biol. 2010;20:811–817. doi: 10.1016/j.cub.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R, Silva E, Clayton L, Gardano L, Barrios-Rodiles M, Wrana J, Varelas X, Arbouzova NI, Shaw S, Saburi S, Matakatsu H, Blair S, McNeill H. Phosphorylation of the tumor suppressor fat is regulated by its ligand Dachsous and the kinase discs overgrown. Curr Biol. 2009;19:1112–1117. doi: 10.1016/j.cub.2009.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley BK, Irvine KD. Warts and Yorkie mediate intestinal regeneration by influencing stem cell proliferation. Curr Biol. 2010;20:1580–1587. doi: 10.1016/j.cub.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Irvine KD. Regulation of Hippo signaling by Jun kinase signaling during compensatory cell proliferation and regeneration, and in neoplastic tumors. Developmental biology. 2011;350:139–151. doi: 10.1016/j.ydbio.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber DA, Hariharan IK. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Tepass U, Theres C, Knust E. crumbs encodes an EGF-like protein expressed on apical membranes of Drosophila epithelial cells and required for organization of epithelia. Cell. 1990;61:787–799. doi: 10.1016/0092-8674(90)90189-l. [DOI] [PubMed] [Google Scholar]
- Thompson BJ, Cohen SM. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell. 2006;126:767–774. doi: 10.1016/j.cell.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Tyler DM, Baker NE. Expanded and fat regulate growth and differentiation in the Drosophila eye through multiple signaling pathways. Dev Biol. 2007;305:187–201. doi: 10.1016/j.ydbio.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- Varelas X, Miller BW, Sopko R, Song S, Gregorieff A, Fellouse FA, Sakuma R, Pawson T, Hunziker W, McNeill H, Wrana JL, Attisano L. The Hippo pathway regulates Wnt/beta-catenin signaling. Dev Cell. 2010;18:579–591. doi: 10.1016/j.devcel.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Wang K, Degerny C, Xu M, Yang XJ. YAP, TAZ, and Yorkie: a conserved family of signal-responsive transcriptional coregulators in animal development and human disease. Biochem Cell Biol. 2009;87:77–91. doi: 10.1139/O08-114. [DOI] [PubMed] [Google Scholar]
- Wang Y, Gilmore TD. Zyxin and paxillin proteins: focal adhesion plaque LIM domain proteins go nuclear. Biochim Biophys Acta. 2003;1593:115–120. doi: 10.1016/s0167-4889(02)00349-x. [DOI] [PubMed] [Google Scholar]
- Watson KL, Justice RW, Bryant PJ. Drosophila in cancer research: the first fifty tumor suppressor genes. J Cell Sci Suppl. 1994;18:19–33. doi: 10.1242/jcs.1994.supplement_18.4. [DOI] [PubMed] [Google Scholar]
- Wei X, Shimizu T, Lai ZC. Mob as tumor suppressor is activated by Hippo kinase for growth inhibition in Drosophila. EMBO J. 2007;26:1772–1781. doi: 10.1038/sj.emboj.7601630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao C, Zhang X, Halder G. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr Biol. 2006;16:2090–2100. doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Willecke M, Hamaratoglu F, Sansores-Garcia L, Tao C, Halder G. Boundaries of Dachsous Cadherin activity modulate the Hippo signaling pathway to induce cell proliferation. Proc Natl Acad Sci U S A. 2008;105:14897–14902. doi: 10.1073/pnas.0805201105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell. 2008;14:388–398. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- Yan Y, Denef N, Tang C, Schupbach T. Drosophila PI4KIIIalpha is required in follicle cells for oocyte polarization and Hippo signaling. Development. 2011;138:1697–1703. doi: 10.1242/dev.059279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Poulton J, Huang YC, Deng WM. The hippo pathway promotes Notch signaling in regulation of cell differentiation, proliferation, and oocyte polarity. PLoS One. 2008;3:e1761. doi: 10.1371/journal.pone.0001761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Zheng Y, Dong J, Klusza S, Deng WM, Pan D. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev Cell. 2010;18:288–299. doi: 10.1016/j.devcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecca M, Struhl G. A feed-forward circuit linking wingless, fat-dachsous signaling, and the warts-hippo pathway to Drosophila wing growth. PLoS Biol. 2010;8:e1000386. doi: 10.1371/journal.pbio.1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ji JY, Yu M, Overholtzer M, Smolen GA, Wang R, Brugge JS, Dyson NJ, Haber DA. YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo pathway. Nat Cell Biol. 2009a;11:1444–1450. doi: 10.1038/ncb1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell. 2008;14:377–387. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Milton CC, Humbert PO, Harvey KF. Transcriptional output of the Salvador/warts/hippo pathway is controlled in distinct fashions in Drosophila melanogaster and mammalian cell lines. Cancer Res. 2009b;69:6033–6041. doi: 10.1158/0008-5472.CAN-08-4592. [DOI] [PubMed] [Google Scholar]
- Zhang X, Milton CC, Poon CL, Hong W, Harvey KF. Wbp2 cooperates with Yorkie to drive tissue growth downstream of the Salvador-Warts-Hippo pathway. Cell Death Differ. 2011 doi: 10.1038/cdd.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, Zheng P, Ye K, Chinnaiyan A, Halder G, Lai ZC, Guan KL. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Szafranski P, Hall CA, Goode S. Basolateral junctions utilize warts signaling to control epithelial-mesenchymal transition and proliferation crucial for migration and invasion of Drosophila ovarian epithelial cells. Genetics. 2008;178:1947–1971. doi: 10.1534/genetics.108.086983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziosi M, Baena-Lopez LA, Grifoni D, Froldi F, Pession A, Garoia F, Trotta V, Bellosta P, Cavicchi S. dMyc functions downstream of Yorkie to promote the supercompetitive behavior of hippo pathway mutant cells. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001140. [DOI] [PMC free article] [PubMed] [Google Scholar]