Abstract
The ancient Egyptians mummified an abundance of cats during the Late Period (664 - 332 BC). The overlapping morphology and sizes of developing wildcats and domestic cats confounds the identity of mummified cat species. Genetic analyses should support mummy identification and was conducted on two long bones and a mandible of three cats that were mummified by the ancient Egyptians. The mummy DNA was extracted in a dedicated ancient DNA laboratory at the University of California – Davis, then directly sequencing between 246 and 402 bp of the mtDNA control region from each bone. When compared to a dataset of wildcats (Felis silvestris silvestris, F. s. tristrami, and F. chaus) as well as a previously published worldwide dataset of modern domestic cat samples, including Egypt, the DNA evidence suggests the three mummies represent common contemporary domestic cat mitotypes prevalent in modern Egypt and the Middle East. Divergence estimates date the origin of the mummies’ mitotypes to between two and 7.5 thousand years prior to their mummification, likely prior to or during Egyptian Predyanstic and Early Dynastic Periods. These data are the first genetic evidence supporting that the ancient Egyptians used domesticated cats, F. s. catus, for votive mummies, and likely implies cats were domesticated prior to extensive mummification of cats.
Keywords: ancient DNA, Felis silvestris catus, mitochondrial, control region, domestication
1. Introduction
Ancient Egyptian culture is well known for its reverence and mummification of cats (Ginsburg, et al., 1991). Cats featured in early Egyptian art and skeletal remains from c. 4000 BC, has led scholars to conclude that our current feline companions might have been domesticated in Egypt (Baldwin, 1975, Ginsburg, et al., 1991, Linseele, et al., 2007). However, the first documentation of wildcat taming, the precursor to domestication, is an archeological finding in Cyprus of a potential wildcat buried with a human, dating to approximately 9,500 years ago (Vigne, et al., 2004), implying prior to the Predynastic Period in Egypt. Recent genetic studies have suggested that the origins of cat domestication occurred in the adjacent Near Eastern sites (Driscoll, et al., 2007, Lipinski, et al., 2008) as domestic cats have derived mitotypes from regional wildcats and the genetic diversity of modern domestic cats is highest within these regions. The cats in Egypt might well have originated from the Near East, perhaps migrating to Egypt via the Levant with trade as already domesticated animals.
The significance of cats to the ancient Egyptians is richly manifested in their ubiquity in tomb art and statuary, and as manifestation of the goddess of beauty, Bastet, or certain aspects of the sun god, Re. Animal mummification was a long-standing tradition in Egypt, reaching its zenith during the Late Period (664 - 332 BC), and continuing through the Ptolemaic and Roman Periods (332 B.C. – AD 395) (Ikram, 2005a). Animal mummies in Egypt can be divided into four categories: pets, revered gods, food offerings and votive offerings (Ikram, 2003, Ikram, 2005a). In their role as totemic animals of Bastet, votive cat mummies were purchased by worshiping pilgrims, offered at temples, and then buried in catacombs (Ikram, 2005a, Zivie and Lichtenberg, 2005). Hence, the majority of mummies found in Egypt and in museum collections are of the votive type. As archeological and permafrost specimens of small wildcats and precursors to domestic cats are not abundant, the votive cat mummies are currently the best source for dating the cat domestication process.
Literally millions of cat mummies were offered and buried in areas sacred to Bastet throughout Egypt. Radiography of votive cat mummies has shown that a majority met their end by unnatural means, mostly due to spinal dislocation or cranial fracturing (Zivie and Lichtenberg, 2005). To supply the demand for votive offerings, catteries were established to raise large numbers of felines for slaughter (Malek, 2006). Cats of all ages and species, perhaps obtained through collection, were given as offerings. Based on morphometric analyses, some felid mummified remains are attributed to the varied subspecies of F. silvestris (Armitage and Clutton-Brock, 1981, Ikram, et al., 2002, Neer and Linseele, 2002) and F. chaus (Armitage and Clutton-Brock, 1981, Ikram, et al., 2002), two small wild felids that co-habited the Nile River Valley region during the reign of the pharaohs, and are still present in a small number today (Osborn and Helmy, 1980). However, the morphometric analyses of cat mummy caches are not always conclusive in identifying the species due to the overlapping sizes and growth metrics of the commensal felids. Indeed, morphometric studies of modern specimens of Felis s. catus, the domesticated cat, and several wildcat subspecies, including F. s. silvestris, the European wildcat, and F. s. tristrami (a.k.a F. libyca tristrami) the Arabian or Middle Eastern wildcat, and F. chaus, Jungle cat, suggest cat species and subspecies are difficult to decipher (Kratochvil, 1975, Randi, et al., 2001, Yamaguchi, et al., 2004). As all these species have been purported to exist in ancient Egypt, genetic analysis must be used to conclusively determine the species of felid contained within cat mummies.
In this study, a portion of the mitochondrial DNA (mtDNA) of three cat mummies was sequenced to decipher the species of cat mummified and the genetic relationship between ancient Egyptian and modern day domestic cats. Because the cat has approximately 50% of its mtDNA genome transposed to numerous regions in the nuclear DNA (numt) (Antunes, et al., 2007, Lopez, et al., 1996), variation from a generated sequence of numt regions can be difficult to define as true mtDNA variation, numt DNA variation, or possibly contamination from modern cat mtDNA. Thus, the mtDNA control region (CR) was selected for analysis as it is only found in the mitochondrial genome and has been shown to contain a significant amount of variability (Grahn, et al., 2011, Tarditi, et al., 2011). In addition, a sufficient dataset exists that allowed the development of mtDNA CR primers in areas of conservation across a majority of mitotypes and also supported comparison to modern cat populations from the Mediterranean, Egypt, the Near East and other regions of the world (Grahn, et al., 2011, Tarditi, et al., 2011).
2. Materials and methods
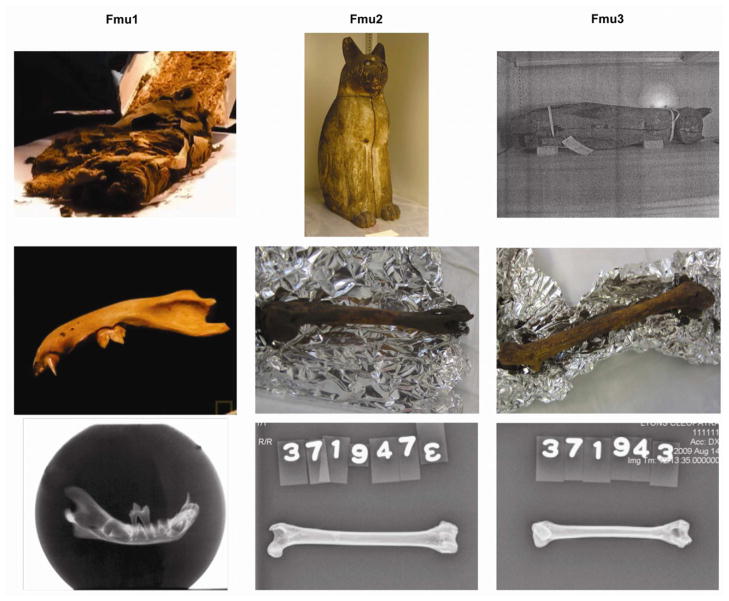
Three cat mummy samples were available for analysis (Table 1, Fig. 1). A mandible from a juvenile cat that was found within cat mummy wrappings was provided by the Hearst Museum of Anthropology in Berkeley, California (Fmu1) (Fig. 1). The sample age and location of origin is unknown, although tentatively estimated to c. 400 BC - 200 AD (Ikram, personal communication). Two additional samples were supplied by the Brooklyn Museum in New York, New York. Both Brooklyn Museum samples were long weight bearing bones, a femur and a humerus, which were extracted from intact mummies (Fmu2 and Fmu3, respectively). The mummies were encased in wooden coffins that depicted the cats in a sitting position with the front paws together (Fig. 1). Both samples were estimated to be from the Late Period of ancient Egypt to the Ptolemaic and Romans Periods between 664 - 332 B.C. The exact provenance of the cat mummies has not been documented. Modern wildcat DNA samples from a previous study (Lipinski, et al., 2008) representing F. chaus (n = 2), F. s. silvestris (n = 22) from and F.s. tristrami (n = 4) from Israel were also analyzed, however in a different laboratory.
Table 1.
Mummified cat sample information and sequencing results.
| Mummy | Museum | Accession No. | Bone | Est. Date* | Sequence | Mitotype |
|---|---|---|---|---|---|---|
| Fmu1 | Phoebe A. Hearst | Mandible | 400 BC – 200 AD | 399 bp | G | |
| Fmu2 | Brooklyn | 37.1943E | Femur | 664 - 332 BC | 402 bp | CB/B2/B3/B5/D/D2/D3/D5/J |
| Fmu3 | Brooklyn | 37.1947E | Humerus | 664 - 332 BC | 246 bp |
Egyptian origins are unknown for the mummies.
Unknown dating however estimated by S. Ikram.
Figure 1. Mummified cat bones for mtDNA CR analysis.
Top) Primary encasement, Middle) Right mandible, left femur, left humerus, Bottom) radiological images. Fmu1 was supplied by the Phoebe A. Hearst Museum of Anthropology in Berkeley, CA, and Fmu2 and Fmu3 from the collection of the Brooklyn Museum, Brooklyn, NY.
A dedicated laboratory space was used for all ancient sample preparations, DNA extractions, and PCR reactions. This ancient DNA laboratory is an isolated building with an independent HEPA-filtered air source. Amplified PCR product has never been transferred into this laboratory. All laboratory equipment is periodically sterilized with bleach and DNA Away wipes (Molecular BioProducts, San Diego, CA). Additionally, sterile personal protective equipment is used at all times including full isolation gowns, gloves, goggles, facemasks, hair coverings, and shoe coverings. No acquired materials were opened in other laboratory facilities.
Although sterile handling techniques have been used once the samples were in the possession of the laboratory, due to the large numbers of cat mummies, historically commonplace desecration of the tombs, and only recent appreciation for the potential of animal remains to contribute to science, the handling practices employed prior to sample acquisition are impossible to verify or predict. Therefore, the surface of the cat mummy samples was assumed to be heavily contaminated with human DNA. Prior to DNA extraction, the samples were washed with 100% bleach, sodium hypochlorite, for 15 minutes followed by rinsing with sterile water. The outside layer of the bone was then scraped off using a sterile scalpel to access the interior of the bone.
Between 100 and 200 mg of bone was ground using a standard mortar and pestle. DNA extraction procedures were identical to previous described methods of Rohland & Hofreiter (2007), including the later addendum of a reduction in volume of the L2 binding buffer to increase yield (Rohland, et al., 2010). A negative control was created with each extraction to verify the purity of all reagents. The pH was adjusted for the binding step as the standard mummification procedures in ancient Egypt resulted in an excessively alkaline corpse.
Following DNA isolation, the extracts were treated with E. coli DNA ligase in an attempt to seal DNA nicks prior to the denaturing effects of PCR (Pusch, et al., 1998). The ligation was performed as follows: in each 50 μl of DNA extraction was also 1 × E. coli ligase buffer and 10 U of E. coli DNA ligase (Invitrogen Corporation, Carlsbad, CA). This was incubated at 16°C for one hour.
To compare to the previous work by Grahn et al. (2011), the partial mtDNA control region (CR), positions 16814 – 206, was directly sequenced in the ancient samples using a set of three overlapping cat specific primers, CCR1, CCR2 and CCR3, that sequenced the same 402 bp region, however using smaller fragments. The primer pairs were designed to generate fragments of 198, 198 and 241 bp respectively (Supporting Information Table 1, Supporting Information Fig. 1). Each 20 μl PCR reaction contained 5 μl of DNA extract (of varying concentrations), 2.0 mM MgCl2, 1 × PCR buffer II (Applied Biosystems, Foster City, CA), 0.2 mM dNTPs (Denville, Saint-Laurent, Canada), 0.4 mM bovine serum albumin (Sigma, St. Louis, MO), 1.0 μM of each primer (Operon, Huntsville, AL) (Supporting Information Table 1) and 0.125 U of Amplitaq Gold (Applied Biosystems, Foster City, CA). Each reaction was amplified under the following cycling conditions in an MJ Research DNA engine (MJ Research, Waltham, MA): 95 °C for 2 min followed by 60 cycles of 45 s at 95 °C, 45 s at 58 °C, and 45 s at 72 °C. The reaction had a final 30 min extension at 72 °C and PCR products were stored at 4 °C. With every PCR set, two negative controls were included; one a negative control from the DNA extraction step to support verification purity of the extraction and ligation techniques, and the second to support verification of the purity of the PCR reagents. The region was amplified in the modern wildcat samples as in Grahn et al. (20110 with identical primers and conditions.
PCR amplification products, 5 μl, were size separated for 90 min at 90 V on 2% TAE agarose gels. Products were photo-documented on an Alpha imager (Alpha Innotek Corp., San Leandro, CA). Products were purified for subsequent sequencing using ExoSap-IT exonuclease cleanup (USB, Cleveland, OH) according to the manufacturer’s specifications. Each sequencing reaction contained 50 ng of purified PCR product, 1 × sequencing buffer, 0.5 μl of BigDye v3.1 (Applied Biosystems, Foster City, CA) and 0.25 μM of either the forward or reverse amplification primer in a 20 μl reaction. Each reaction was amplified on an MJ Research DNA engine (MJ Research, Waltham, MA) according to the manufacturers specifications with cycles increased from 30 to 40. Unincorporated, labeled nucleotides were removed from the reaction using Sephadex G-50 (Sigma–Aldrich, St. Louis, MO) in a 96-well plate. Sequencing product, approximately 12 μl, was combined with 10 μl of Hi-DI formamide (Applied Biosystems, Foster City, CA) and denatured for 3 min at 95 °C. Products were separated on an ABI 3730 DNA Analyzer.
Sequences were aligned using Sequencher 4.1™ software (Gene Codes Corporation, Ann Arbor, MI). Sequencing data were assembled into contigs and then aligned manually to the Sylvester Reference Sequence (SRS) (Grahn, et al., 2011) using the program BioEdit v. 7.0.9.0 (Hall, 1999) and matched to previously identified mitotypes representing 1,394 global F. s. catus samples (Grahn, et al., 2011, Tarditi, et al., 2011). Samples were also compared to an additional 446 F. s. silvestris sequences (Personal communication, RA Grahn). Estimates of divergence times were calculated as in Lopez et al. (1997) and ranges were calibrated using both the neutral and fast calibrations.
3. Results
Full mitotype sequences were obtained for 28 modern wildcat samples with the exception of a single individual for which the final seven base pairs could not be determined. Five new mitotypes were discovered (Genbank JQ245443-JQ245447, Supporting Information Fig. 2), novel mutations discovered within the wildcat species are summarized in Table 2. In this dataset, each wildcat maintained species-specific mutations: F. s. tristrami had a deletion of base 262 from the SRS, F. s. silvestris consistently presented a diagnostic 2 bp deletion of SRS 367–368, and F. chaus had 12 mutations not seen in any other felid species to date. However, in the expanded dataset of 446 F. s. silvestris sequences, many wildcats share mitotypes common to domestic cats (unpublished data).
Table 2.
Cat mummy and wildcat mitotype defining nucleotides.
| Lopez | SRS | NTP | Fmu1 | Fmu2 | Fmu3 | Wildcat Mitotypes
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| U20753 | Pos. | U | C | B/D/J 1 | Fc | Fst-A | Fst-B | Fss-A | Fss-B | |
| 16820 | 7 | T | . | . | . | C | C | C | C | C |
| 16822 | 9 | C | . | . | . | G | . | . | . | . |
| 16824 | 11 | A | . | G | G | . | . | . | . | . |
| 16834 | 21 | T | . | . | . | . | . | . | C | C |
| 16852 | 39 | C | . | . | . | T | . | . | . | . |
| 16859 | 46 | C | . | T | . | T | T | T | T | T |
| 16867 | 54 | C | . | . | . | T | . | . | . | . |
| 16899 | 86 | T | . | . | . | . | . | . | C | C |
| 16918.1 | 105.1 | – | . | . | . | T | . | . | . | . |
| 16931 | 118 | C | . | . | . | T | . | . | . | . |
| 16956 | 143 | G | . | . | . | A | . | . | . | . |
| 16961 | 148 | A | . | . | . | . | . | . | G | G |
| 16962 | 149 | A | . | . | . | . | . | . | G | G |
| 16963 | 150 | A | . | . | . | G | . | . | . | . |
| 16966 | 153 | C | . | . | . | T | . | . | . | T |
| 16970 | 157 | G | . | . | . | A | . | . | . | . |
| 16985 | 172 | A | . | G | . | . | . | . | G | G |
| 16986 | 173 | T | . | . | . | C | . | . | . | . |
| 16988 | 175 | C | . | . | . | . | T | T | . | . |
| 16997 | 184 | G | . | . | . | . | A | A | A | A |
| 59 | 255 | C | T | T | . | . | . | . | . | |
| 62.1 | 259 | – | . | . | . | T | T | T | T | |
| 63 | 260 | T | A | A | . | A | A | A | A | |
| 70 | 267 | A | . | . | C | . | . | . | . | |
| 75 | 272 | G | . | . | . | – | – | . | . | |
| 75.1 | 272.1 | – | . | . | G | . | . | . | . | |
| 80 | 277 | A | . | . | G | . | . | . | . | |
| 131 | 328 | T | . | . | . | . | . | A | . | |
| 140 | 337 | G | . | . | A | . | . | . | . | |
| 159 | 356 | T | C | C | . | . | . | C | C | |
| 168 | 365 | G | . | . | . | . | . | A | A | |
| 169 | 366 | T | . | . | . | . | . | C | C | |
| 170 | 367 | A | . | . | . | . | . | – | – | |
| 171 | 368 | C | . | . | . | . | . | – | – | |
| 173 | 370 | G | . | . | – | . | . | A | A | |
| 196 | 393 | A | . | . | . | . | C | . | . | |
|
| ||||||||||
| n = 2 | n = 3 | n = 1 | n = 13 | n = 9 | ||||||
Mitotype defining sites are indicated based on the first published sequence (Lopez et al., 1996) and the SRS (Sylvester reference sequence) (Grahn et al., 2011). FC, F. chaus; FLT, F. lybica tristrami; FSS, F. silvestris silvestris.
Fmu3 may also be the following mitotypes: B2/B3/B5/D2/D3/D5
As expected from previous ancient DNA experiments, PCR amplification of some DNA fragments were sporadic in the mummies (Pääbo, et al., 2004). Eleven amplification attempts produced results. Primer pair CCR1 was successfully amplified once in each mummy. Primer pair CCR2 was successfully amplified twice in mummies Fmu1 and Fmul 2. The third primer set, CCR3, successfully amplified Fmu1 twice, once in Fmu2, but failed in Fmu3. Successful amplification and sequencing replicates matched with 100% correspondence. Additionally, sequence data were verified by overlapping regions from independent amplifications. However, only forward sequence was obtained for CCR2 and CCR3 in Fmu1.
A complete 402 bp sequence was generated for Fmu2. Fmu1 failed to produce quality sequence in only a three bp segment, producing 399 bp of sequence. As primer set CCR3 failed to amplify in sample Fmu3, only 246 bp sequence was generated. Comparison to published cat CR mtDNA sequences supported the determination of the mummy mitotypes (Grahn, et al., 2011, Tarditi, et al., 2011). Despite incomplete data, the mitotype of Fmu1 and Fmu2 was identified, and the mitotype of Fmu3 was restricted to a family of mitotypes. Fmu1 appears to be a derivative of domestic cat mitotype G but contains a SNP that makes it unique among all samples and sequences evaluated. Fmu2 represents mitotype C and Fmu3 may belong to mitotypes B, D, J, or one of their closely related mitotypes (Table 2, Supporting Information Fig. 2). None of the mummified samples contained any diagnostic mutations indicative of a wild felid, although Fmu3 cannot conclusively be determined (Table 2, Supporting Information Fig. 2). Divergence calculations estimate that Fmu1 and Fmu2 diverged between 1.99 and 7.5 thousand years before their existence, and Fmu3 diverged from the other two between 1.97 and 4.1 thousand years prior to their birth.
4. Discussion
Both the scientific and lay communities are fascinated with Egyptian mummies. The most common association of mummies is with the ancient pharaohs, however, a variety of animals were mummified as votive offerings, particularly cats, the vast quantity of which suggest Egyptians may then have been the first to domesticate cats. The first physical evidence of potentially domesticated cats in Egypt consists of the skeletal remains found in a tomb at Mostagedda (c. 4000 BC) and one at Abydos (c. 3000 BC), followed by a handful of textual mentions of cats in the Old Kingdom (c. 2663 - 2195 BC). During the Middle Kingdom (c. 2066 - 1650 BC), cats become more common, both in text and images, including in contexts that are clearly indicative of domestication (for example, Neferronpet Kenro, c. 1250 BC Theban Tomb 178).
The first indication of cat mummification is dated to c. 1350 B.C., as represented by an elaborately carved limestone sarcophagus, and is assumed to have contained Prince Thutmose’s beloved pet (Ikram, et al., 2002). Animal mummies were prepared in a variety of methods by the ancient Egyptians, the most common being desiccation and anointment. Each step of the votive mummification process is highly antagonistic to the chemistry of PCR (Ikram, 2005b), including desiccating the body with natron, a naturally occurring Egyptian salt (Na2CO3•10H2O), anointing the body with heated oils and resins, wrapping in linen, and then sometimes pouring additional hot resins and/or oil over the body (Ikram, 2005b). Following the aforementioned treatments, votive mummies were kept in large, collective tombs or catacombs. Although the majority of the cat mummy caches are mostly located in the desert and are dry, occasionally the tombs were flooded, both in antiquity and during modern times. The combinations of high heat, high humidity, extreme alkaline conditions, and sometimes fire caused by spontaneous combustion (Zivie and Lichtenberg, 2005) have spurred debate as to the feasibility of ancient DNA studies on Egyptian remains (Gilbert, et al., 2005, Zink and Nerlich, 2005).
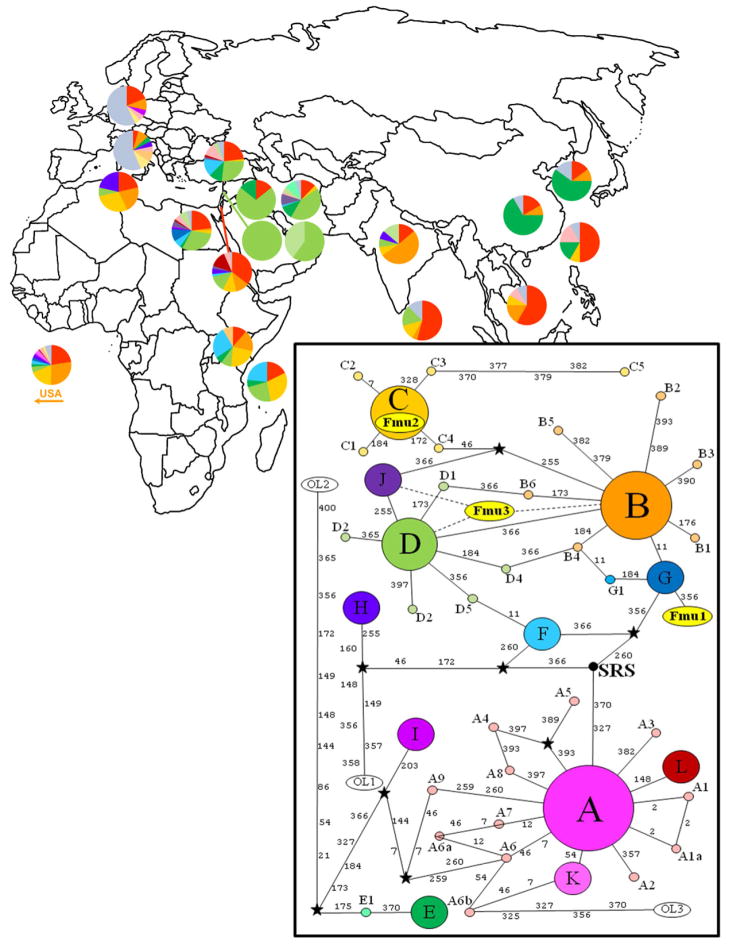
Despite the unfavorable treatment and conditions of mummification, the analysis of at least mtDNA, which is found in higher copy numbers than nuclear DNA, has proven feasible for cat mummy investigations. While inconsistent success may be due to mutations in the priming regions, failure in the PCR is most likely due to a paucity of intact mitochondrial fragments. Mitochondrial DNA sequence was obtained from each of the three cat mummies, including two weight-bearing bones and a mandible. Exact mtDNA CR mitotypes could be interpreted for two of the cat mummies and a subset of mitotypes was implicated for the third mummy. Fmu1 had a unique mitotype when compared to over 1800 domestic and wildcat sequences from the feline mtDNA CR. Mummy Fmu1’s type is one mutation removed from mitotype G, a mitotype that is found in 10% of modern Egyptian cats and at a very low frequency in the United States, perhaps due to recent importation of cats from Egypt. The unique mitotypes supports, at least in this mummy, that the sequence data does not represent contemporary contamination. Cat mummy Fmu2 had mitotype C, which is present in 3% of modern Egyptian cats and 12% worldwide. Data, albeit incomplete, suggested the third mummy belongs to mitotype B, sub-types B2, B3, or B5, mitotype D, sub-types D2, D3, D5, or mitotype J. Mitotype B is common in the Middle East and Indian Ocean trade routes and represents 1.5% of modern Egyptian cats; Mitotype D represents approximately 30% of modern Egyptian cats and 13% of the world’s cats and is predominantly in the Mediterranean and Middle East, and mitotype J is only found in Egypt and the Middle East (Grahn, et al., 2011). All three mummies are representative of closely related mitotypes that are still present in the modern Egyptian population. All three mitoypes are part of the same clade as found by Grahn et al. (2011); a deep division separating mitotypes B/C/D/F/G/J from mitotypes A/E/I/K/L. Furthermore, the mitotypes were of types rare to Western Europe and the United States, but common to the Middle East, particularly Egypt, indicating that modern cats of Egypt are descendents of local ancient populations.
A prediction of wild felid species of the mummies was attempted using the mtDNA CR sequences. As neither the two base pair deletion common in F. s. silvestris from Switzerland, nor the insertions present in F. chaus or F. s. tristrami were found in Fmu1 or Fmu2, suggesting the mummies are not wild felids. The diagnostic sites could not be assayed in Fmu3. However, a large dataset has been recently generated, yet unpublished, that indicates that most European wildcats have mitotypes shared with domestic cats (RA Grahn, personal communication). In addition, other sub-species of wildcats should be examined for a more thorough comparison. Divergence calculations estimates of the mitotypes are based on the Lopez model (1997), which were calibrated based on mutation rates of the 12s and ND2 genes within the mitochondria, one having a faster and one having a lower mutation rate, respectively. The coalescence of the mitotypes estimates that the common ancestor of the mummies existed between 1.97 and 7.5 thousand years before their existence, supporting cat domestication to have occurred during the time of agricultural development in the Middle East, likely slight prior to or during the Predynastic and Early Dynastic Periods of Egypt.
Cat domestication is likely to have occurred in the Near East, including the Fertile Crescent regions (Driscoll, et al., 2007, Lipinski, et al., 2008, Vigne, et al., 2004), however, only recently were modern Egyptian cats considered in expanded analyses (Kurushima, Submitted). Contacts between ancient Egypt, the Eastern Mediterranean and the Near East are well attested from 3500 B.C. and even earlier. Thus, domestic (or tamed) cats could have entered Egypt from northern regions as early as the Predynastic Period. By the era of extensive mummifications from the Late Period through the advent of Christianity, cats would have been abundant and heterogeneous. The three cat mummies with three different mitotypes is indicative of high mtDNA diversity within the domestic cat population 2,500 years ago, suggesting that large and diverse domestic cat populations predate the era when mummified votive offerings were common.
A great burden to ancient DNA researchers is proof of legitimacy of the results. While criteria for ensuring authenticity has been suggested by many (Cooper and Poinar, 2000, Pääbo, et al., 2004, Richards, et al., 1995, Yang and Watt, 2005), few studies can follow all of the suggestions (Roberts and Ingham, 2008). This study attempted to follow as many of the aforementioned criteria as possible, including: 1) the use of an isolated and sterile laboratory, 2) cleaning of samples with an external sterilization, using bleach as well as removal of the surface layers, 3) numerous positive and negative controls were implemented in every procedure, and 4) multiple replicates were performed with both the extraction procedures and the sequencing protocols. Other potential problems, such as the unknown sterility of the excavation conditions and potential modern human contamination, were avoided through thorough removal of the external layers of the samples and the use of Felis specific mtDNA primers. In addition, the significant overlap of sequence assisted the verification of authentic DNA sequences by selectively targeting cat DNA to eliminate human contamination of PCR products, as well as acting as internal replicates to verify that the DNA is, in fact, authentic mummified cat. These replicates reduce the chance of miss-interpretation of single base mutations, as suggested by Pääbo et al. (2004). Thus, cat mummy studies should have a higher legitimacy and less concern for human contamination as compared to the Neanderthal and King Tutankhamun analyses (Cooper and Poinar, 2000, Hawass, et al., 2010, Kimura, et al., 2011, Yang and Watt, 2005).
This study marks the first successful amplification of DNA from Egyptian cat mummies. The mitotypes of the cat mummies still exist in the present day populations, allowing modern cats to trace their genealogy to the time of the Pharaohs. Although the first steps in cat domestication might have occurred in Cyprus and the Near East (Driscoll, et al., 2007, Lipinski, et al., 2008, Vigne, et al., 2004), the Egyptians may well have been the first cat breeders, both of which were important steps in the domestication process of cats. Continued DNA studies of the extensive collections of mummified cats should decipher the domestication history of cats as seen in Egypt’s Late Period.
Supplementary Material
Figure 2. Distribution and mitotype network of modern worldwide cat populations.
Fmu1 is mitotype G. Fmu2 is mitotype C. Fmu3 is mitotype D, or derived mitotype J. All mummy mitotypes represent a grouping distinct from the most common mitotype A (adapted from Grahn et al., 2011). Pie charts represent the percentage of each mitotype found at each location by Grahn et al., 2011. Gray color (as seen predominantly in Germany and Italy) represents the unique mitotypes.
Highlights.
mtDNA is extracted and sequenced from Egyptian cat mummies.
Ancient Egyptian cat mummies are domestic cats, and not wild species.
Each cat mummy had a different mitotype currently found in modern Egyptian feral cat populations.
Cats were domesticated prior to or during the Predynastic or Early Dynastic Periods of Egypt.
Acknowledgments
Partial funding was provided by the National Institutes of Health – National Center for Research Resources (R24 RR016094) and is currently supported by the Office of Research Infrastructure Programs/OD R24OD010928 and a National Geographic Expedition Grant (EC0360-07) (LAL). Financial support was supplied by the University of California – Davis, Veterinary Genetics Laboratory and the Center for Companion Animal Health for the establishment of a dedicated animal ancient DNA laboratory space. We appreciate the donation of wildcat samples from Drs. Hans Lutz and Alan Levy. We appreciate the assistance of Dr. Bernie May with construction of the ancient DNA laboratory. We appreciate Lisa Bruno of the Brooklyn Museum in New York, NY, for her expertise, and Drs. Robert K. Wayne and Klaus-Peter Koepfli for training in ancient DNA laboratory techniques at their facilities at the University of California, Los Angeles.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antunes A, Pontius J, Ramos MJ, O’Brien SJ, Johnson WE. Mitochondrial introgressions into the nuclear genome of the domestic cat. Journal of Heredity. 2007;98:414–420. doi: 10.1093/jhered/esm062. [DOI] [PubMed] [Google Scholar]
- Armitage PL, Clutton-Brock J. A radiological and histological investigation into the mummification of cats from Ancient Egypt. Journal of Archaeological Science. 1981;8:185–196. [Google Scholar]
- Baldwin JA. Notes and Speculations on Domestication of Cat in Egypt. Anthropos. 1975;70:428–448. [Google Scholar]
- Cooper A, Poinar HN. Ancient DNA: Do It Right or Not at All. Science. 2000;289:1139. doi: 10.1126/science.289.5482.1139b. [DOI] [PubMed] [Google Scholar]
- Driscoll CA, Menotti-Raymond M, Roca AL, Hupe K, Johnson WE, Geffen E, Harley EH, Delibes M, Pontier D, Kitchener AC, Yamaguchi N, O’Brien SJ, Macdonald DW. The Near Eastern Origin of Cat Domestication. Science. 2007;317:519–523. doi: 10.1126/science.1139518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MTP, Barnes I, Collins MJ, Smith C, Eklund J, Goudsmit J, Poinar H, Cooper A. Long-term survival of ancient DNA in Egypt: Response to Zink and Nerlich (2003) American Journal of Physical Anthropology. 2005;128:110–114. doi: 10.1002/ajpa.20045. [DOI] [PubMed] [Google Scholar]
- Ginsburg L, Delibrias G, Minaut-Gout A, Valladas H, Zivie A. On the Egyptian Origin of the Domestic Cat. Bulletin du Museum National d’Histoire Naturelle Section C Sciences de la Terre Paleontologie Geologie Mineralogie. 1991;13:107–114. [Google Scholar]
- Grahn RA, Kurushima JD, Billings NC, Grahn JC, Halverson JL, Hammer E, Ho CK, Kun TJ, Levy JK, Lipinski MJ, Mwenda JM, Ozpinar H, Schuster RK, Shoorijeh SJ, Tarditi CR, Waly NE, Wictum EJ, Lyons LA. Feline non-repetitive mitochondrial DNA control region database for forensic evidence. Forensic Science International: Genetics. 2011;5:33–42. doi: 10.1016/j.fsigen.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Hawass Z, Gad YZ, Ismail S, Khairat R, Fathalla D, Hasan N, Ahmed A, Elleithy H, Ball M, Gaballah F, Wasef S, Fateen M, Amer H, Gostner P, Selim A, Zink A, Pusch CM. Ancestry and Pathology in King Tutankhamun’s Family. JAMA-Journal of the American Medical Association. 2010;303:638–647. doi: 10.1001/jama.2010.121. [DOI] [PubMed] [Google Scholar]
- Ikram S. Death and burial in ancient Egypt. Longman; Harlow: 2003. [Google Scholar]
- Ikram S. Divine creatures : animal mummies in ancient Egypt. American University in Cairo Press; Cairo; New York: 2005a. [Google Scholar]
- Ikram S. Manufacturing Divinity: The Technology of Mummification. In: Ikram S, editor. Divine Creatures: Animal Mummies in Ancient Egypt. The American University in Cairo Press; Cairo: 2005b. pp. 16–43. [Google Scholar]
- Ikram S, Iskander N, Egypt Wizarat a-T, Majlis al-A’la l-A, Mathaf a-M. Catalogue general of Egyptian antiquities in the Cairo Museum : nos. 24048–24056; 29504–29903 (selected); 51084–51101; 61089 : non-human mummies. Supreme Council of Antiquities Press; Cairo: 2002. [Google Scholar]
- Kimura B, Marshall FB, Chen SY, Rosenbom S, Moehlman PD, Tuross N, Sabin RC, Peters J, Barich B, Yohannes H, Kebede F, Teclai R, Beja-Pereira A, Mulligan CJ. Ancient DNA from Nubian and Somali wild ass provides insights into donkey ancestry and domestication. Proceedings of the Royal Society B-Biological Sciences. 2011;278:50–57. doi: 10.1098/rspb.2010.0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratochvil Z. The Value Relationships of Pairs of Skull Characteristics as Taxonomic Criteria for Felis-Silvestris-Silvestris and Felis-Silvestris-F-Catus Mammalia. Zoologicke Listy. 1975;24:13–19. [Google Scholar]
- Linseele V, Van Neer W, Hendrickx S. Evidence for early cat taming in Egypt. Journal of Archaeological Science. 2007;34:2081–2090. [Google Scholar]
- Lipinski MJ, Froenicke L, Baysac KC, Billings NC, Leutenegger CM, Levy AM, Longeri M, Niini T, Ozpinar H, Slater MR, Pedersen NC, Lyons LA. The ascent of cat breeds: Genetic evaluations of breeds and worldwide random-bred populations. Genomics. 2008;91:12–21. doi: 10.1016/j.ygeno.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez JV, Cevario S, O’Brien SJ. Complete Nucleotide Sequences of the Domestic Cat (Felis catus) Mitochondrial Genome and a Transposed mtDNA Tandem Repeat (Numt) in the Nuclear Genome. Genomics. 1996;33:229–246. doi: 10.1006/geno.1996.0188. [DOI] [PubMed] [Google Scholar]
- Lopez JV, Culver M, Stephens JC, Johnson WE, Obrien SJ. Rates of nuclear and cytoplasmic mitochondrial DNA sequence divergence in mammals. Molecular Biology and Evolution. 1997;14:277–286. doi: 10.1093/oxfordjournals.molbev.a025763. [DOI] [PubMed] [Google Scholar]
- Malek J. The cat in ancient Egypt. British Museum Press; London: 2006. [Google Scholar]
- Neer WV, Linseele V. New Analyses of Old Bones: the Faunal Remains from Hierakonpolis. Nekhen News. 2002;14:7–8. [Google Scholar]
- Osborn DJ, Helmy I. The contemporary land mammals of Egypt (including Sinai) Field Museum of Natural History; Chicago, Ill: 1980. [Google Scholar]
- Pääbo S, Poinar H, Serre D, Jaenicke-Despres V, Hebler J, Rohland N, Kuch M, Krause J, Vigilant L, Hofreiter M. Genetic analyses from ancient DNA. Annual Review of Genetics. 2004;38:645–679. doi: 10.1146/annurev.genet.37.110801.143214. [DOI] [PubMed] [Google Scholar]
- Pusch CM, Giddings I, Scholz M. Repair of degraded duplex DNA from prehistoric samples using Escherichia coli DNA polymerase I and T4 DNA ligase. Nucleic Acids Research. 1998;26:857–859. doi: 10.1093/nar/26.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randi E, Pierpaoli M, Beaumont M, Ragni B, Sforzi A. Genetic identification of wild and domestic cats (Felis silvestris) and their hybrids using Bayesian clustering methods. Molecular Biology and Evolution. 2001;18:1679–1693. doi: 10.1093/oxfordjournals.molbev.a003956. [DOI] [PubMed] [Google Scholar]
- Richards MB, Sykes BC, Hedges REM. Authenticating DNA Extracted from Ancient Skeletal Remains. Journal of Archaeological Science. 1995;22:291–299. [Google Scholar]
- Roberts C, Ingham S. Using Ancient DNA Analysis in Palaeopathology: A Critical Analysis of Published Papers, with Recommendations for Future Work. International Journal of Osteoarchaeology. 2008;18:600–613. [Google Scholar]
- Rohland N, Hofreiter M. Comparison and optimization of ancient DNA extraction. Biotechniques. 2007;42:343–352. doi: 10.2144/000112383. [DOI] [PubMed] [Google Scholar]
- Rohland N, Siedel H, Hofreiter M. A rapid column-based ancient DNA extraction method for increased sample throughput. Molecular Ecology Resources. 2010;10:677–683. doi: 10.1111/j.1755-0998.2009.02824.x. [DOI] [PubMed] [Google Scholar]
- Tarditi CR, Grahn RA, Evans JJ, Kurushima JD, Lyons LA. Mitochondrial DNA Sequencing of Cat Hair: An Informative Forensic Tool. Journal of Forensic Sciences. 2011;56:S36–S46. doi: 10.1111/j.1556-4029.2010.01592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigne JD, Guilaine J, Debue K, Haye L, Gerard P. Early taming of the cat in Cyprus. Science. 2004;304:259–259. doi: 10.1126/science.1095335. [DOI] [PubMed] [Google Scholar]
- Yamaguchi N, Driscoll CA, Kitchener AC, Ward JM, Macdonald DW. Craniological differentiation between European wildcats (Felis silvestris silvestris), African wildcats (F. s. lybica) and Asian wildcats (F. s. ornata): implications for their evolution and conservation. Biological Journal of the Linnean Society. 2004;83:47–63. [Google Scholar]
- Yang DY, Watt K. Contamination controls when preparing archaeological remains for ancient DNA analysis. Journal of Archaeological Science. 2005;32:331–336. [Google Scholar]
- Zink AR, Nerlich AG. Long-term survival of ancient DNA in Egypt: Reply to Gilbert et al. American Journal of Physical Anthropology. 2005;128:115–118. doi: 10.1002/ajpa.20045. [DOI] [PubMed] [Google Scholar]
- Zivie A, Lichtenberg R. The Cats of the Goddess Bastet. In: Ikram S, editor. Divine Creatures: Animal Mummies in Ancient Egypt. The American University in Cairo Press; Cairo: 2005. pp. 106–119. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.