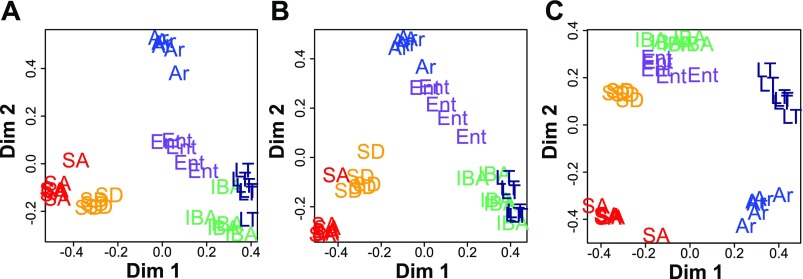
Fig. 3.
Random forests (RF) clustering of individual samples based on protein spot abundance. The results of RF analysis using 418 (of 2,073) protein spots that were matched on all gels (A), the 89 (of 150) protein spots with unique IDs that were present on all gels (B), and the top 7 spots from the analysis in B (C). These proteins, which most strongly separate individuals from different physiological states and cluster those within states, are apolipoprotein A-I (APOA1, 2 spots), mitofilin (IMMT), dihydrolipoamide S-succinyltransferase (DLST), aspartoacylase (ASPA), albumin (ALB), and guanine nucleotide binding protein (GNB2). These proteins are indicated in Fig. 2. Stages are as defined in Fig. 1 with n = 5 for SD, IBA, and Ent, and n = 6 for SA, LT, and Ar.