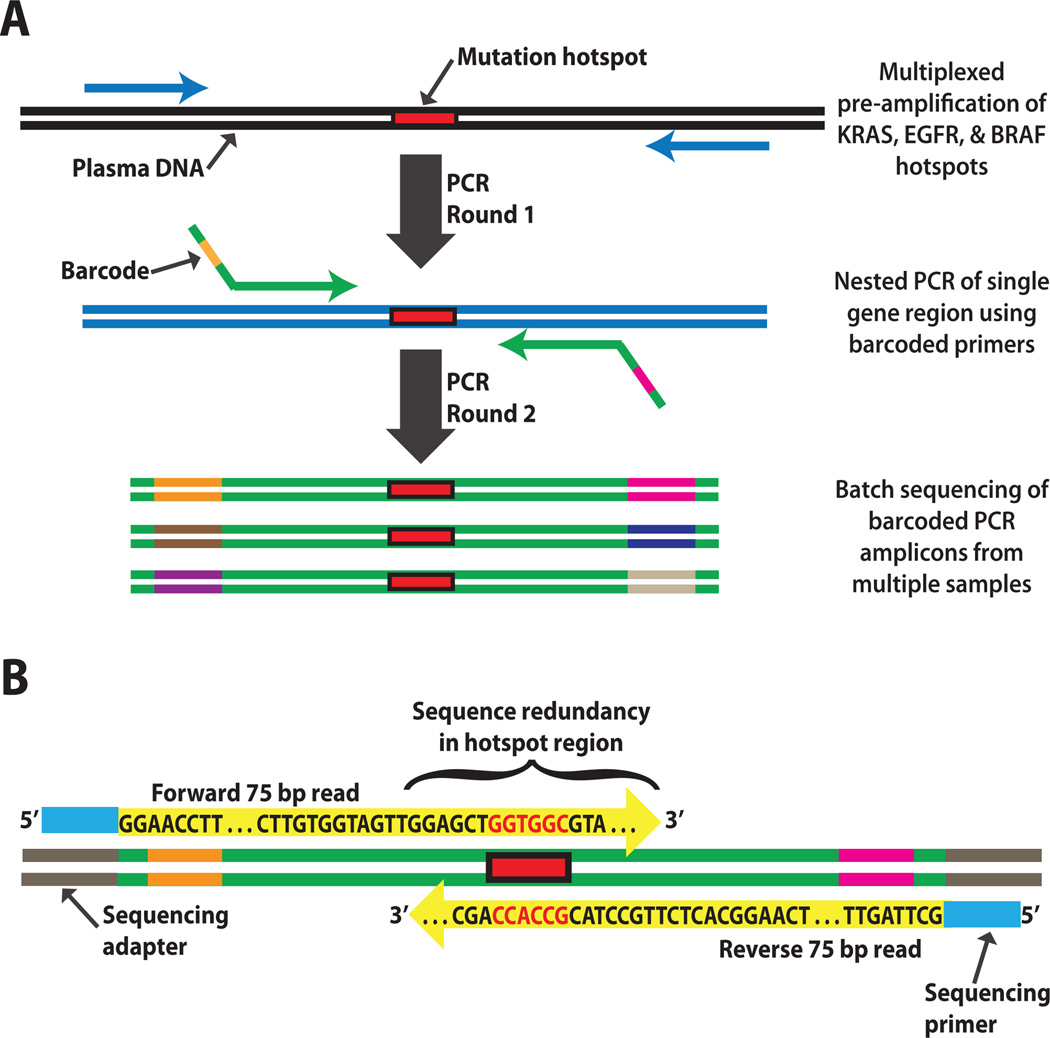
Fig. 1. Schematic of the error-suppressed multiplexed deep sequencing approach.
(A) Cell-free DNA purified from plasma undergoes two rounds of amplification by PCR. The first round amplifies mutation hotspot regions of several genes from a given sample in a single tube. The second round separately amplifies each hotspot region using nested primers incorporating unique combinations of barcodes to label distinct samples. The barcoded PCR products are then pooled and subjected to deep sequencing. Millions of sequences are sorted and counted to determine the ratio of mutant to wild-type molecules derived from each sample. The total number of plasma DNA fragments is measured by real-time PCR and can be used to calculate the absolute concentration of mutant ctDNA. (B) Sequence redundancy in mutation hotspot regions is produced by partial overlap of paired-end reads from the forward and reverse strands of each clone. This yields highly accurate base-calls, permitting detection and quantitation of rare mutations with greater sensitivity.