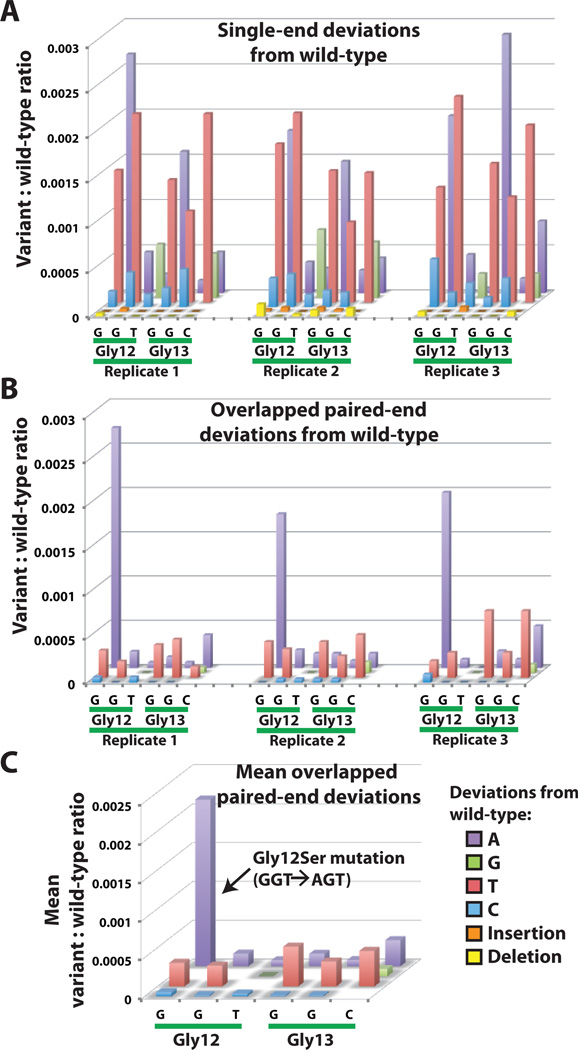
Fig. 2. Suppression of spurious mutation counts to reveal low-abundance variants.
Each bar indicates the frequency of a particular deviation from the wild-type sequence occurring within the codon 12/13 hotspot region of KRAS. The tested sample contains 0.2% DNA derived from a lung cancer cell line that is known to be homozygous for a KRAS Gly12Ser mutation. (A) Filtered reads from one end of the amplicon have relatively frequent mismatches when directly compared to the wild-type sequence. Data from 3 replicate amplifications are shown. (B) Sequencer errors are greatly reduced by requiring both partially overlapping paired-end reads from each clone to exactly match a specific mutation. The Gly12Ser mutation is now readily distinguished from the remaining low-level errors that were likely introduced during DNA amplification and processing. Insertions and deletions are no longer seen in this region after requiring agreement of overlapped reads. (C) A further reduction in the relative error level can be achieved by calculating the mean values of 3 replicate measurements, since mutations found in the original DNA sample should produce more consistent counts than randomly occurring errors.