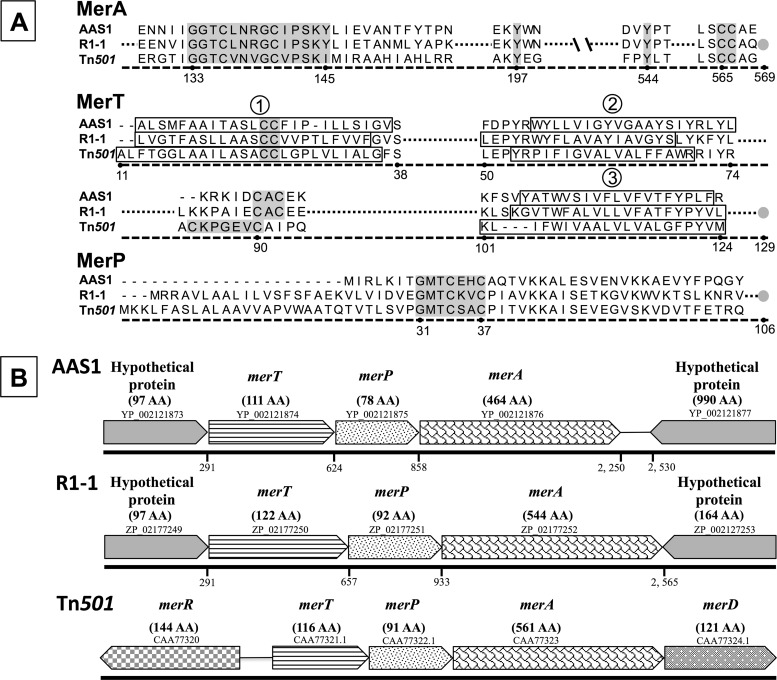
Fig 1.
(A) Alignment of putative Mer proteins, including MerA, MerT, and MerP from Hydrogenobaculum sp. Y04AAS1 (AAS1) and Hydrogenivirga sp. 128-5-R1-1 (R1-1), with mer sequences from Pseudomonas aeruginosa Tn501 as a reference. Highlighted conserved regions of functional importance (3) are as follows: for MerA, the redox active site, two downstream tyrosines, and the carboxy terminus-vicinal CC pair; for MerT, the conserved C residues in the first membrane-spanning domain and in the cytoplasmic loop between the second and third membrane-spanning domains; and for MerP, the metal binding motif GMTCxxC. The membrane-spanning domains (determined using TMpred [http://www.ch.embnet.org/software/TMPRED_form.html]) are boxed (numbered 1 to 3) in MerT. Gray circles indicate the carboxy termini of the proteins. Numbering indicates position in the Tn501 sequence. (B) mer operon gene order. Arrowed boxes indicate each ORF and the direction of transcription; accession numbers are included above each box. Names of putative gene products and corresponding numbers of amino acids (AA) are given above the boxes. The numbered line below each ORF represents the nucleotide position marking the start of the gene counted from the transcription start nucleotide upstream of the hypothetical proteins in each operon.