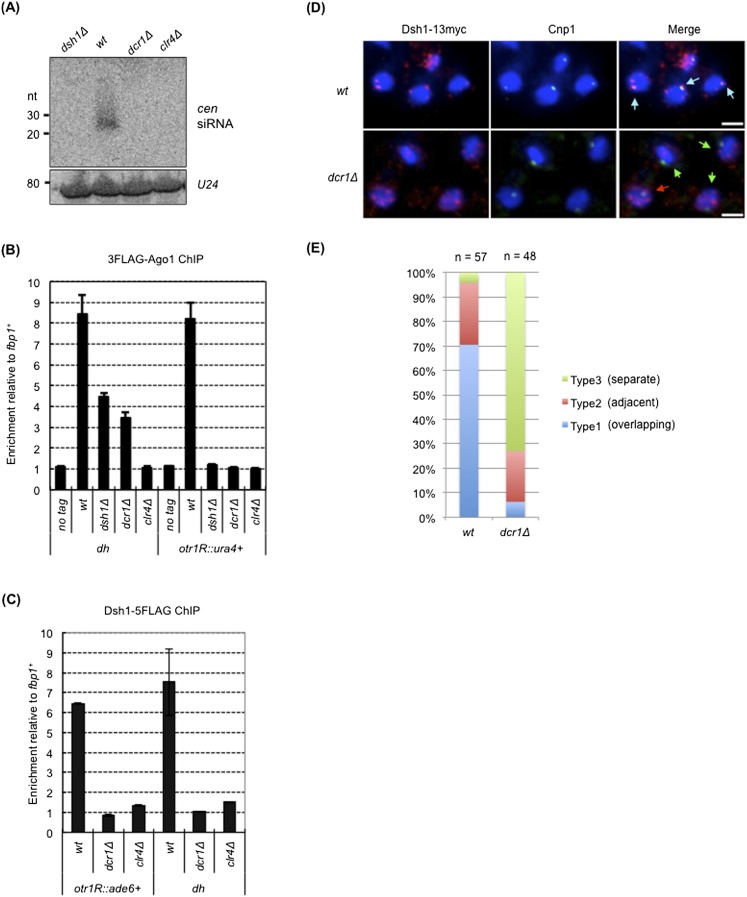
Figure 2.
dsh1+ is an essential component of the RNAi machinery. (A) siRNA analysis by Northern blotting using radiolabeled oligonucleotide probes against dg and dh centromeric repeats. A probe against U24 RNA was also used as an internal control. (B) ChIP analysis of 3×Flag-Ago1 at otr1R∷ura4+ and dh in the indicated strains. Enrichment relative to fbp1+ is shown on the Y-axis. (C) ChIP analysis of Dsh1-5Flag at otr1R∷ade6+ and dh. dcr1Δ and clr4Δ mutants were used as controls for RNAi mutants and heterochromatin mutants, respectively. Enrichment relative to fbp1+ is shown on the Y-axis. (D) Indirect immunofluorescence analysis of Dsh1 localization in a strain expressing Dsh1-13myc. Cells growing exponentially were stained with DAPI, an antibody against myc, and an antibody against Cnp1 to visualize DNA, Dsh1-13myc, and the position of the centromeres, respectively. White bars, 2 μm. (E) The fraction of cells showing overlapping, adjacent, and separate signals of Dsh1-13myc and Cnp1 (indicated by blue, red, and green arrows, respectively, in the right panels in D) in the cells showing both signals are indicated by a bar graph.