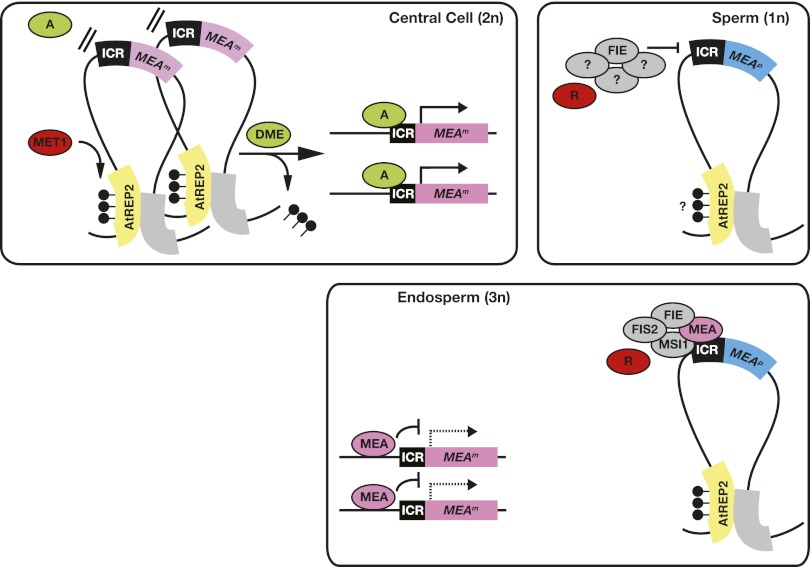
Figure 6.
Model of MEA imprinting control through a higher-order chromatin structure. Methylation at AtREP2 is maintained by MET1 in central cells and sperm cells. AtREP2 might interact with another region, thereby forming a repressive chromatin loop preventing the MEA locus from being accessed by a transcriptional activator (A). Specific expression of DME in the central cell removes methylation and resolves the repressive chromatin loop. This allows the transcriptional activator (A) to access the MEA-ICR. As a consequence, in the endosperm, the two maternal MEA alleles (MEAm) are expressed. Paternal MEA allele (MEAp) silencing is maintained by a proposed PcG complex containing FIE during male gametogenesis (Jullien et al. 2006a). After fertilization, MEAp is repressed partially by the maternal MEA–FIE complex and another paternal repressor (R). Since parental alleles in the endosperm are differentially targeted by trans-acting factors, they must have been marked in the germline, as illustrated by the purple and blue color of the maternal and paternal allele, respectively. The nature of this mark is unknown. The model explicitly shows MEAm activation in the central cell for imprinted expression in the endosperm; however, the same model is proposed for the egg cell and embryo. (Lollipops) DNA methylation; (dashed line) autorepressed MEAm transcription.