Abstract
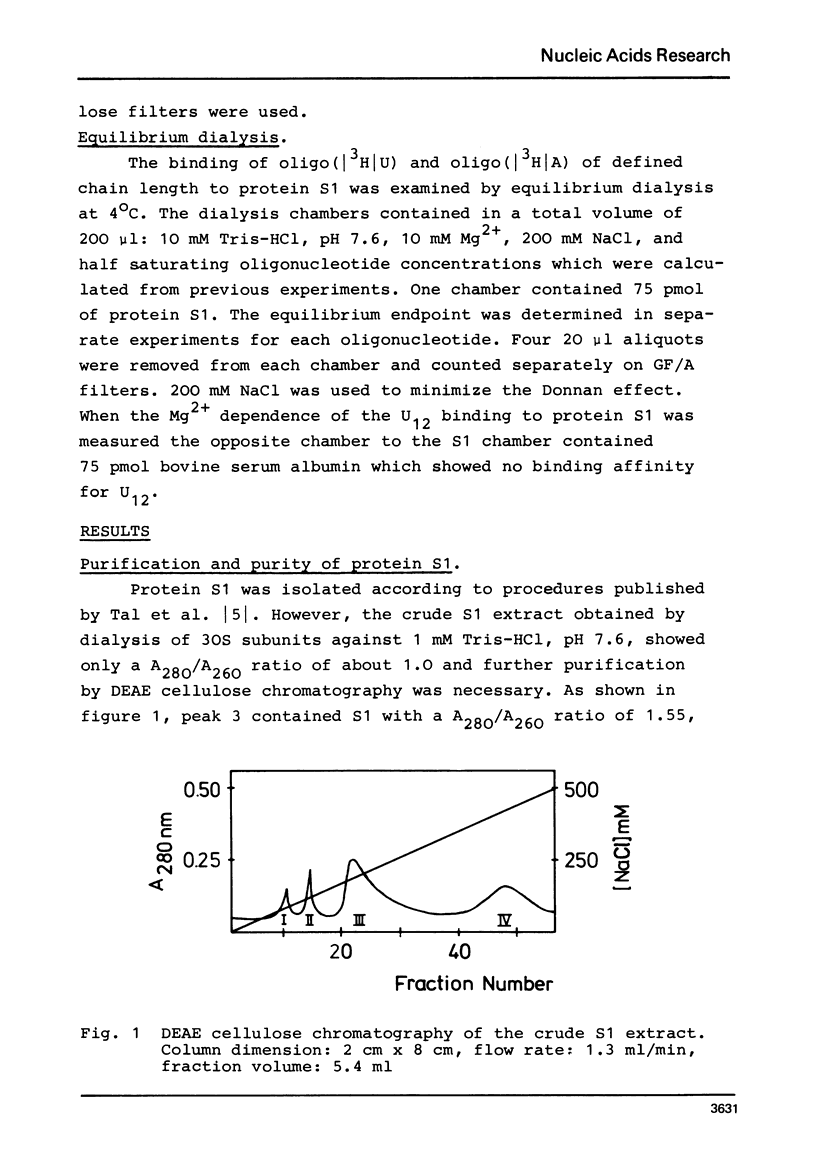
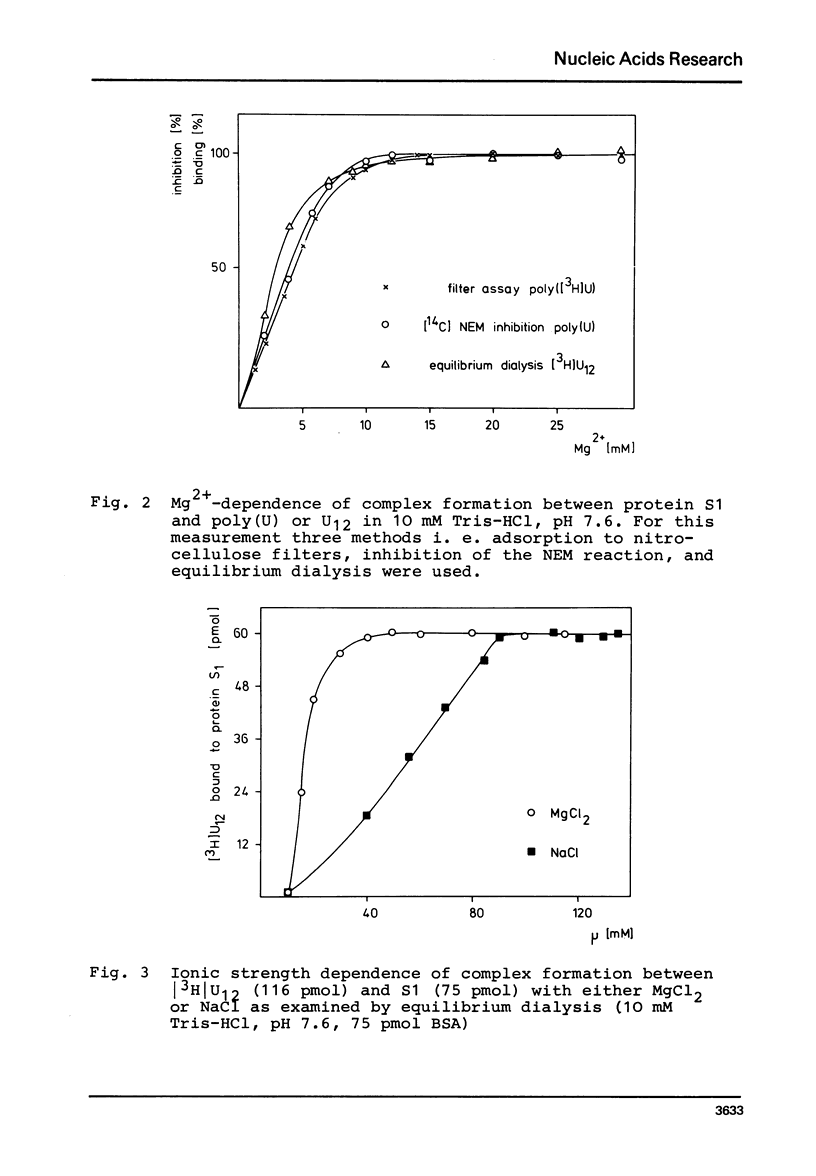
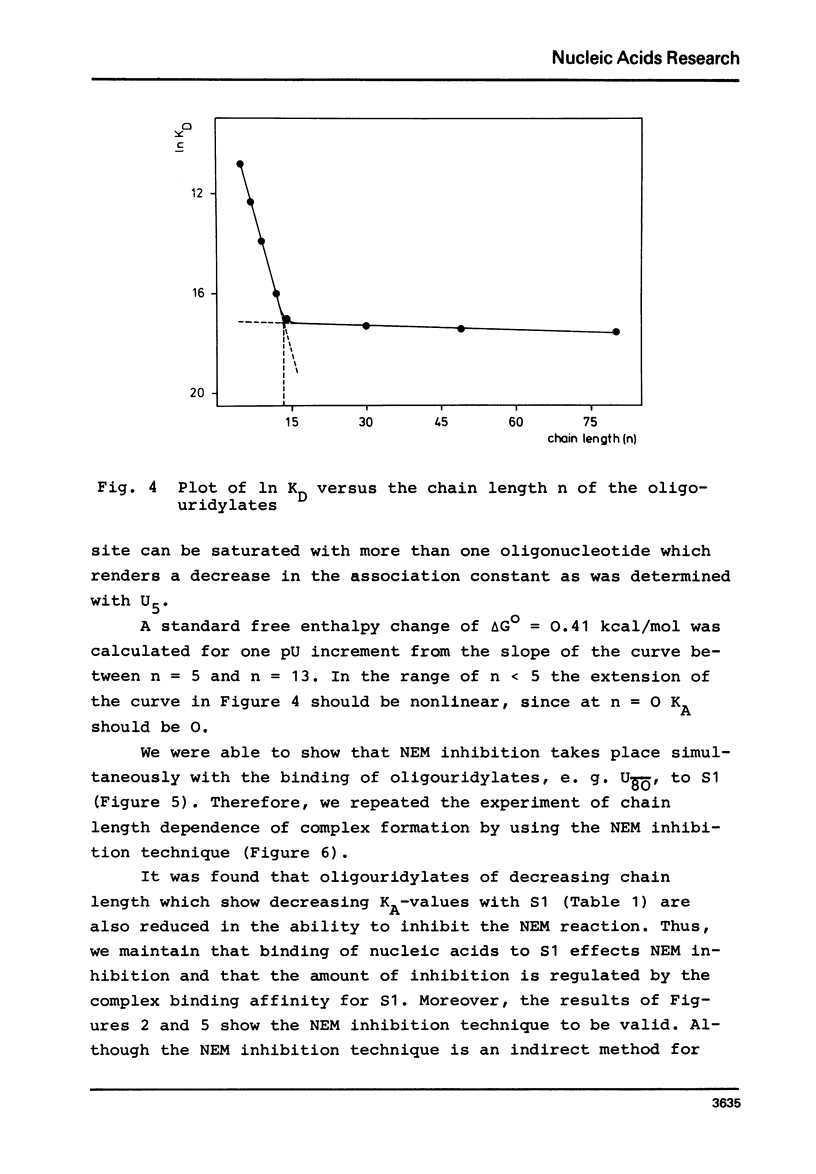
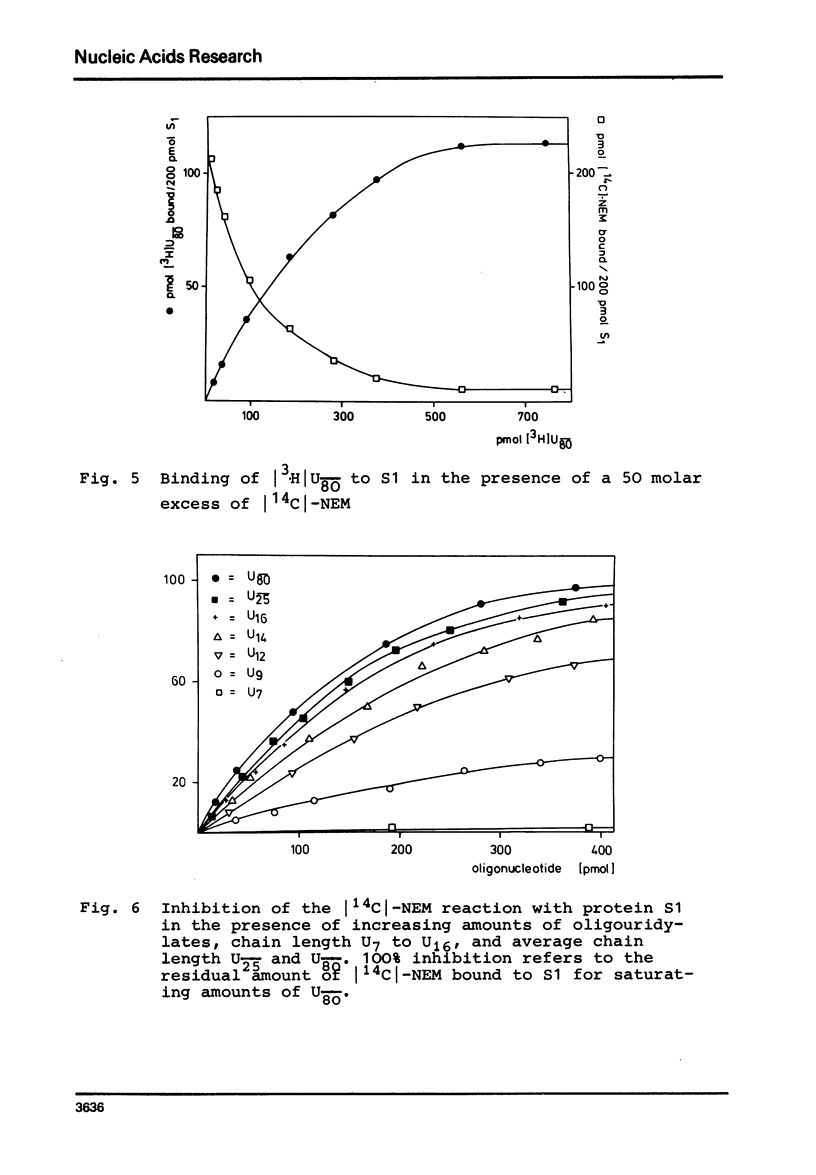
In order to examine the nature of the complex formation between the ribosomal protein S1 and nucleic acids three methods were used: Inhibition of the reaction of n-ethyl[2.3 14C]-maleimide with S1 by the addition of oligonucleotides; adsorption of the complexes to nitrocellulose filters; and equilibrium dialysis. The complex formation is Mg2+ dependent at low salt concentrations and becomes Mg2+ independent at an ionic strength greater than 90 mM. Oligouridylates of increasing chain length reach an optimal KA of 3-3-10(7) M-1 at a chain length of n=13-14. Protein S1 contains one binding site for long chain oligouridylates, such as U12, and the standard-free-energy change on binding caused by one Pu increment is 0.41 kcal/mol, when n varies between five and fourteen. Complex formation is insensitive to the capacity of the homopolynucleotide bases to form hydrogen bonds. Homopolynuceotides, however, showing a Tm less than 250 in the buffer system used show an increased affinity for S1 compared to poly(A) and poly(C) (Tm greater than 40 degrees). The data are discussed with respect to the proposed binding of protein S1 to the 3-terminal end of the 16S RNA.
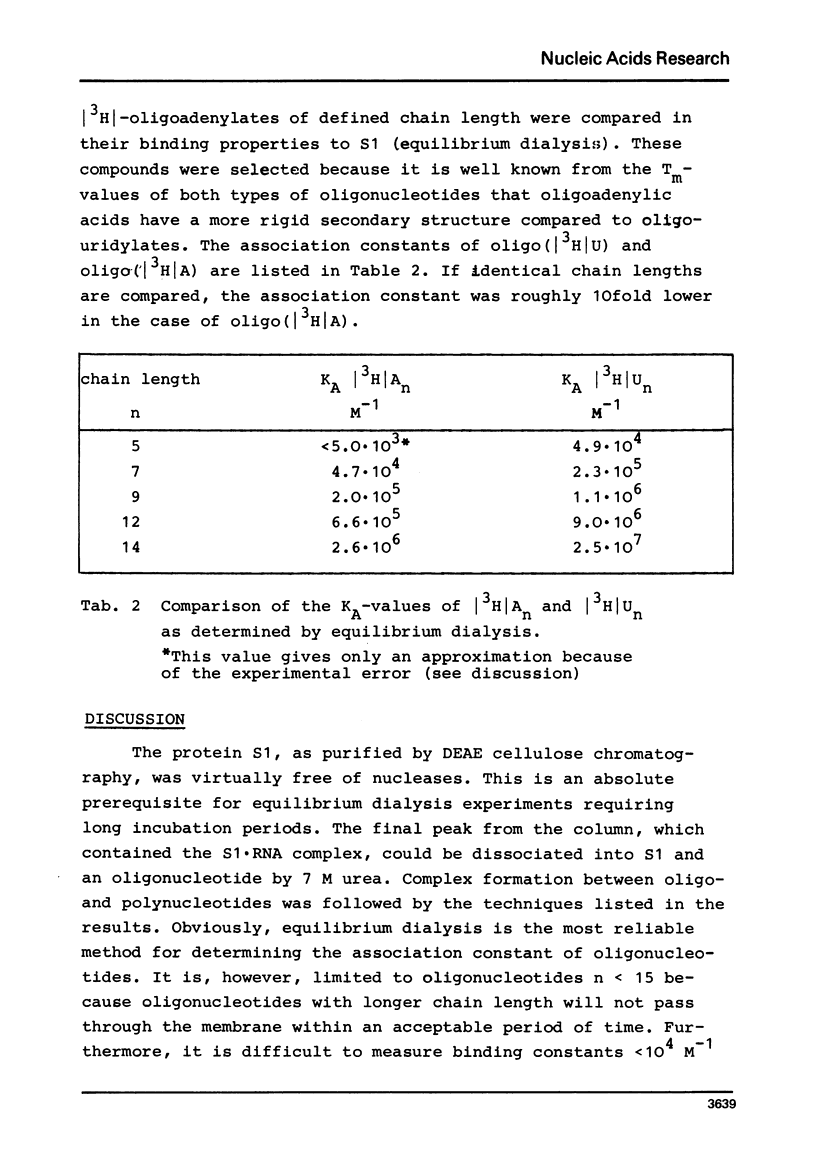
Full text
PDF









Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baan R. A., Hilbers C. W., Van Charldorp R., Van Leerdam E., Van Knippenberg P. H., Bosch L. High-resolution proton magnetic resonance study of the secondary structure of the 3'-terminal 49-nucleotide fragment of 16S rRNA from Escherichia coli. Proc Natl Acad Sci U S A. 1977 Mar;74(3):1028–1031. doi: 10.1073/pnas.74.3.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear D. G., Ng R., Van Derveer D., Johnson N. P., Thomas G., Schleich T., Noller H. F. Alteration of polynucleotide secondary structure by ribosomal protein S1. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1824–1828. doi: 10.1073/pnas.73.6.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
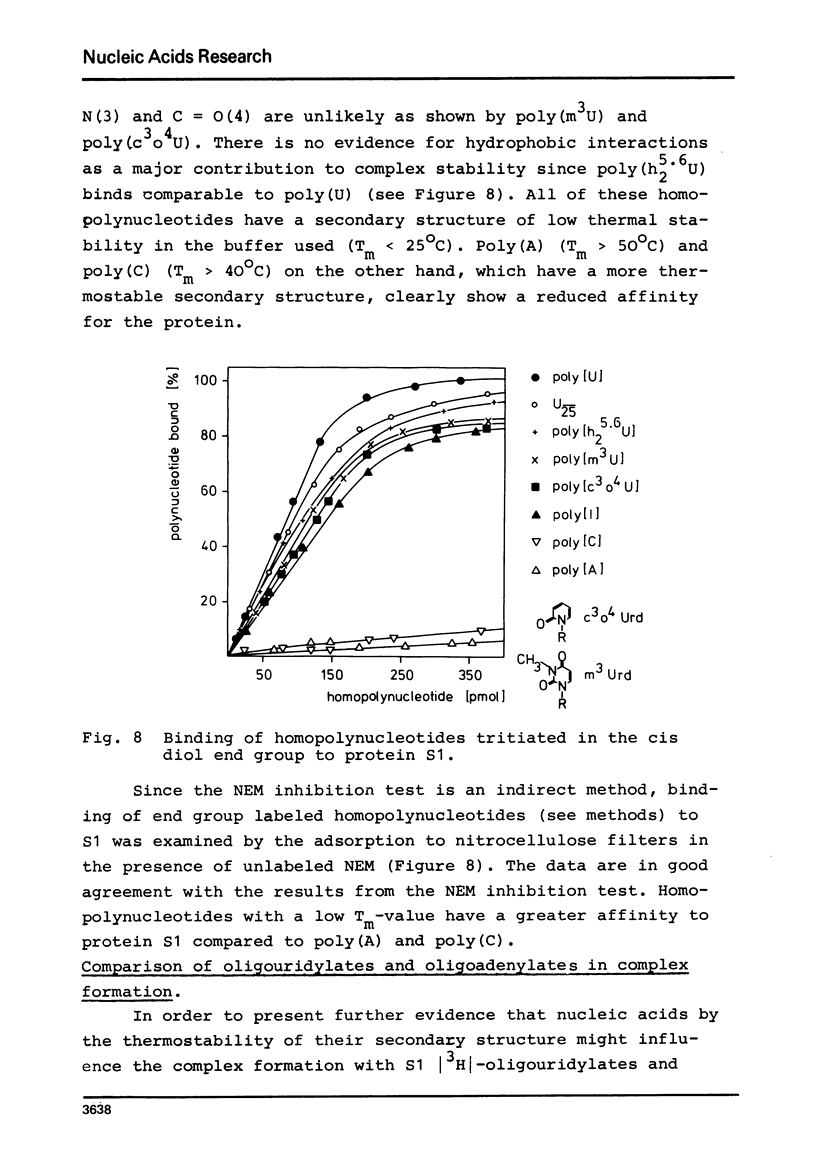
- CERUTTI P., IKEDA K., WITKOP B. THE SELECTIVE PHOTOREDUCTION OF URIDINE IN POLYNUCLEOTIDES. J Am Chem Soc. 1965 Jun 5;87:2505–2507. doi: 10.1021/ja01089a045. [DOI] [PubMed] [Google Scholar]
- Dahlberg A. E., Dahlberg J. E. Binding of ribosomal protein S1 of Escherichia coli to the 3' end of 16S rRNA. Proc Natl Acad Sci U S A. 1975 Aug;72(8):2940–2944. doi: 10.1073/pnas.72.8.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermoso J. M., Szer W. Replacement of ribosomal protein S1 by interference factor ialpha in ribosomal binding of phage Ms2 RNA. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4708–4712. doi: 10.1073/pnas.71.12.4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamen R., Kondo M., Römer W., Weissmann C. Reconstitution of Q replicase lacking subunit with protein-synthesis-interference factor i. Eur J Biochem. 1972 Nov 21;31(1):44–51. doi: 10.1111/j.1432-1033.1972.tb02498.x. [DOI] [PubMed] [Google Scholar]
- Kolakofsky D., Weissmann C. Possible mechanism for transition of viral RNA from polysome to replication complex. Nat New Biol. 1971 May 12;231(19):42–46. doi: 10.1038/newbio231042a0. [DOI] [PubMed] [Google Scholar]
- Küppers B., Sumper M. Minimal requirements for template recognition by bacteriophage Qbeta replicase: approach to general RNA-dependent RNA synthesis. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2640–2643. doi: 10.1073/pnas.72.7.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughrea M., Moore P. B. Physical properties of ribosomal protein S1 and its interaction with the 30 S ribosomal subunit of Escherichia coli. J Mol Biol. 1977 May 25;112(3):399–421. doi: 10.1016/s0022-2836(77)80189-7. [DOI] [PubMed] [Google Scholar]
- Miller M. J., Niveleau A., Wahba A. J. Inhibition of synthetic and natural messenger translation. I. Purification and properties of a protein isolated from Escherichia coli MRE 600 ribosomes. J Biol Chem. 1974 Jun 25;249(12):3803–3807. [PubMed] [Google Scholar]
- Miller M. J., Wahba A. J. Inhibition of synthetic and natural messenger translation. II. Specificity and mechanism of action of a protein isolated from Escherichia coli MRE 600 ribosomes. J Biol Chem. 1974 Jun 25;249(12):3808–3813. [PubMed] [Google Scholar]
- Moore G., Crichton R. R. Reductive methylation: a method for preparing functionally active radioactive ribosomes. FEBS Lett. 1973 Nov 15;37(1):74–78. doi: 10.1016/0014-5793(73)80429-6. [DOI] [PubMed] [Google Scholar]
- Pochon F., Michelson A. M. Polynucleotides. IX. Methylation of nucleic acids, homopolynucleotides and complexes. Biochim Biophys Acta. 1967 Nov 21;149(1):99–106. doi: 10.1016/0005-2787(67)90694-6. [DOI] [PubMed] [Google Scholar]
- RajBhandary U. L. Studies on polynucleotides. LXXVII. The labeling of end groups in polynucleotide chains: the selective modification of diol end groups in ribonucleic acids. J Biol Chem. 1968 Feb 10;243(3):556–564. [PubMed] [Google Scholar]
- Randerath K., Randerath E., Chia L. S., Gupta R. C., Sivarajan M. Sequence analysis of nonradioactive RNA fragments by periodate-phosphatase digestion and chemical tritium labeling: characterization of large oligonucleotides and oligonucleotides containing modified nucleosides. Nucleic Acids Res. 1974 Sep;1(9):1121–1141. doi: 10.1093/nar/1.9.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schetters H., Gassen H. G., Matthaei H. Codon-anticodon interaction studied with oligonucleotides containing 3 -deazauridine, 4 -deoxyuridine or 3 -deaza- 4 -deoxyuridine. I. Synthesis by primer-dependent polynucleotide phosphorylase of oligonucleotides containing modofied nucleosides. Biochim Biophys Acta. 1972 Jul 31;272(4):549–559. doi: 10.1016/0005-2787(72)90510-2. [DOI] [PubMed] [Google Scholar]
- Schwarz U., Menzel H. M., Gassen H. G. Codon-dependent rearrangement of the three-dimensional structure of phenylalanine tRNA, exposing the T-psi-C-G sequence for binding to the 50S ribosomal subunit. Biochemistry. 1976 Jun 1;15(11):2484–2490. doi: 10.1021/bi00656a035. [DOI] [PubMed] [Google Scholar]
- Senear A. W., Steitz J. A. Site-specific interaction of Qbeta host factor and ribosomal protein S1 with Qbeta and R17 bacteriophage RNAs. J Biol Chem. 1976 Apr 10;251(7):1902–1912. [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolarsky M., Tal M. A protein factor stimulating binding and translating of polyuridylic acid by Escherichia coli ribosomes. Biochim Biophys Acta. 1970 Aug 8;213(2):401–416. doi: 10.1016/0005-2787(70)90048-1. [DOI] [PubMed] [Google Scholar]
- Steitz J. A., Wahba A. J., Laughrea M., Moore P. B. Differential requirements for polypeptide chain initiation complex formation at the three bacteriophage R17 initiator regions. Nucleic Acids Res. 1977 Jan;4(1):1–15. doi: 10.1093/nar/4.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szer W., Hermoso J. M., Boublik M. Destabilization of the secondary structure of RNA by ribosomal protein S1 from Escherichia coli. Biochem Biophys Res Commun. 1976 Jun 7;70(3):957–964. doi: 10.1016/0006-291x(76)90685-9. [DOI] [PubMed] [Google Scholar]
- Tal M., Aviram M., Kanarek A., Weiss A. Polyuridylic acid binding and translating by Escherichia coli ribosomes: stimulation by protein I, inhibition by aurintricarboxylic acid. Biochim Biophys Acta. 1972 Oct 27;281(3):381–392. doi: 10.1016/0005-2787(72)90452-2. [DOI] [PubMed] [Google Scholar]
- Thrierr J. C., Dourlent M., Leng M. A study of polyuridylic acid. J Mol Biol. 1971 Jun 28;58(3):815–830. doi: 10.1016/0022-2836(71)90042-8. [DOI] [PubMed] [Google Scholar]
- Van Dieijen G., Van Der Laken C. J., Van Knippenberg P. H., Van Duin J. Function of Escherichia coli ribosomal protein S1 in translation of natural and synthetic messenger RNA. J Mol Biol. 1975 Apr 15;93(3):351–366. doi: 10.1016/0022-2836(75)90282-x. [DOI] [PubMed] [Google Scholar]
- van Dieijen G., van Knippenberg P. H., van Duin J. The specific role of ribosomal protein S1 in the recognition of native phage RNA. Eur J Biochem. 1976 May 1;64(2):511–518. doi: 10.1111/j.1432-1033.1976.tb10330.x. [DOI] [PubMed] [Google Scholar]


