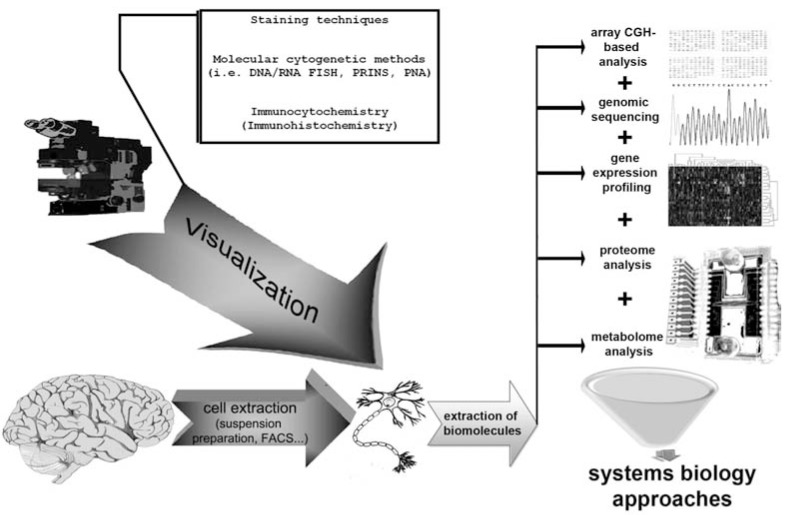
Fig. (1).
Technological principles of single cell genomics of the brain. The first step of any procedure aimed at studying genome/epigenome (proteome/metabolome) at single cell level is cell isolation. The latter can be performed in a variety of manners (i.e. brain cell suspension preparations, FACS or other flow-cytometry-based approaches; for more details see [28, 29]). The obtained cells can be subjected to procedures allowing microscopic visual analysis (visualization) of macromolecules (nucleic acids, proteins etc.) or macromolecular complexes (i.e. chromatin) through direct staining of cells, FISH, immunocytochemistry or immunohistochemistry. Alternatively, extraction of biomolecules can be performed to perform analysis of nucleic acids (DNA/RNA), proteins and metabolites either through on-chip technologies or through mass spectrometry and nuclear magnetic resonance technologies. Moreover, “lab-on-chip” technologies have been recently become available for analyzing simultaneously nucleic acids, proteins and metabolites of a cell [32]. All the data can be processed by systems biology (bioinformatic/in silico) approaches to create an integrated view of genetic, epigenetic, proteomic and metabolomic profiles.