Table 3.
Symbolic representation
| SYMBOL | MEANING |
|---|---|
| D | The gene expression matrix |
| di | ithgene in D |
| δ | Signum threshold |
| G | Co-expression network |
| V | Set of vertices in G |
| E | Set of edges in G |
| Dist | Distance matrix |
| Dist(di, dj) | NMRS distance between genes di, dj∈D |
| Adj | Adjacency matrix |
| Adj(vi,vj) | 1 if vi and vj are connected by an edge 0 otherwise |
| Gcon | Set of connected region |
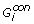 |
ith connected region |
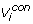 |
Set of vertices in ith connected region |
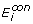 |
Set of edges in ith connected region |
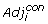 |
Adjacency matrix of the ith connected region |
 |
ith network module |
| Dnet | Set of network modules obtained from G |
| TOM(vi,vj) | Topological Matrix value between vertices vi and vj |
| TOM(V1) | Average TOM of the set of vertices V1 |
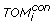 |
TOM for ith connected region |
 |
Maximum spanning tree obtained from ith connected region |
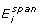 |
Set of edges in 
|
The table 3 describes the various symbols that is used in ModuleMiner.
