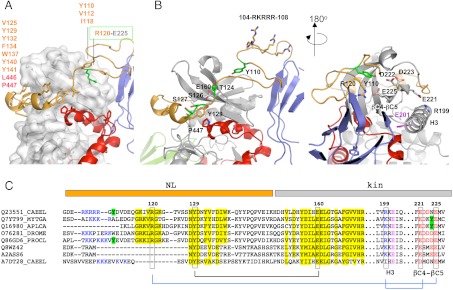
Fig. 2.
The NL flanking region. (A) Hydrophobic groups mediating the packing of the NL against the kinase domain are listed. The buried salt bridge fixing the NL front to the kinase is highlighted; (B) Structural features of the NL segment in two views. Residues shown (or proposed to) undergo phosphorylation are in green. The strictly conserved tyrosine Y129 and its interacting residues are displayed. The cluster of positively charged residues (104–108) is shown. The catalytic glutamate in helix H3 is in magenta; (C) Motif conservation in the NL region. Sequences belong to TwcKs from C. elegans (Q23551), Mytilus galloprovincialis (Q7YT99), and Aplysia californica (Q16980); projectin from Drosophila melanogaster (O76281) and crayfish (Q86GD6); human (Q8WZ42) and mouse (A2ASS6) titin; and TTN-1 from C. elegans (A7DT28). Conservation is highlighted in yellow and interacting residues joined by lines. Positively charged residues at the NL front and negatively charged residues in the βC4-βC5 loop of the N-terminal kinase lobe are blue and red, respectively. Tyrosine residues in this region (or the structurally complementary region in mollusks) are marked in green. The similarity of twitchins and projectins, but not titins or TTN-1, is noticeable. A certain co-variation of the β-hairpin and the βC4-βC5 loop is detected. (Full sequence alignment of twitchins and projectins in Fig. S3).