Abstract
The epigenetic silencing of exogenous transcriptional units integrated into the genome represents a critical problem both for long-term gene therapy efficacy and for the eradication of latent viral infections. We report here that limitation of essential amino acids, such as methionine and cysteine, causes selective up-regulation of exogenous transgene expression in mammalian cells. Prolonged amino acid deprivation led to significant and reversible increase in the expression levels of stably integrated transgenes transcribed by means of viral or human promoters in HeLa cells. This phenomenon was mediated by epigenetic chromatin modifications, because histone deacetylase (HDAC) inhibitors reproduced starvation-induced transgene up-regulation, and transcriptome analysis, ChIP, and pharmacological and RNAi approaches revealed that a specific class II HDAC, namely HDAC4, plays a critical role in maintaining the silencing of exogenous transgenes. This mechanism was also operational in cells chronically infected with HIV-1, the etiological agent of AIDS, in a latency state. Indeed, both amino acid starvation and pharmacological inhibition of HDAC4 promoted reactivation of HIV-1 transcription and reverse transcriptase activity production in HDAC4+ ACH-2 T-lymphocytic cells but not in HDAC4− U1 promonocytic cells. Thus, amino acid deprivation leads to transcriptional derepression of silenced transgenes, including integrated plasmids and retroviruses, by a process involving inactivation or down-regulation of HDAC4. These findings suggest that selective targeting of HDAC4 might represent a unique strategy for modulating the expression of therapeutic viral vectors, as well as that of integrated HIV-1 proviruses in latent reservoirs without significant cytotoxicity.
Keywords: HIV-1 latency, ocular albinism type 1, tyrosine, TNF alpha, GPR143
Mammalian cells respond to deprivation of essential amino acids by a finely tuned response, characterized by profound changes in gene expression and modification of several cellular processes (1), including epigenetic chromatin remodelling, which may lead to long-lasting effects. Together with DNA methylation, histone posttranslational modifications are recognized as major epigenetic marks responsible for transcriptional regulation, and they can be stably transmitted to daughter cells, although they remain reversible (2). Acetylation (i.e., addition of acetyl groups to the ε-amino group of specific lysine residues) is one of the most common modifications of histones, and it is typically associated with transcriptional activation. Indeed, it generates binding sites for bromodomain proteins and neutralizes the positive charge of histones, decreasing their affinity for DNA (3). By contrast, histone deacetylation results in a condensed and transcriptionally repressed chromatin state. The extent of histone acetylation depends on a dynamic process governed by two families of enzymes, termed histone acetyltransferases and histone deacetylases (HDACs), which add and remove acetyl groups from the histone tails, respectively.
HDACs represent a large family of enzymes displaying different structure, function, expression pattern, and subcellular localization, and they are grouped into four classes (3). Class I, II, and IV HDACs are zinc-dependent enzymes, whereas class III HDACs, also referred to as sirtuins (SIRT1–SIRT7), are NAD+-dependent enzymes and constitute a structurally separate family. Class I HDACs include HDAC1, HDAC2, HDAC3, and HDAC8, which are ubiquitously expressed, prevalently localize to the nucleus, and display high deacetylase activity toward histones. Class II HDACs are subdivided into class IIa and class IIb. Class IIa HDAC4, HDAC5, HDAC7, and HDAC9 exhibit a tissue-specific expression pattern, nucleocytoplasmic shuttling, and weak deacetylase activity toward histones in vitro because of the substitution of a highly conserved tyrosine (Y) in the catalytic domain (4); however, their enzymatic activity in vivo could derive from the recruitment of class I HDACs in multiprotein complexes (5). Class IIb HDACs include HDAC6, characterized by cytoplasmic localization and deacetylase activity vs. α-tubulin, and HDAC10, of unknown function. Class IV consists of the poorly characterized HDAC11 only.
Although the precise mode of action and target specificity of this family of enzymes remain to be established, broad-spectrum HDAC inhibitors (HDACi) have been proposed as therapeutic agents in cancer and neuromuscular/neurodegenerative diseases (6–8), as well as in the design of strategies targeting cells chronically infected by HIV-1. In fact, the persistence of long-lived cells latently infected with HIV-1 represents a fundamental barrier to viral eradication in individuals receiving combination antiretroviral therapy (cART) (9). The dormant state of the HIV-1 provirus depends on host transcription factors involved in maintaining an inactive LTR-driven transcription, but also on epigenetic regulation mediated by covalent modifications of histones (10). For this reason, HDACi have been considered in studies of viral reactivation with the objective of purging latently infected cells in individuals assuming cART by rendering them “visible” to the immune system or sensitive to pharmacological treatments (9, 11).
We report here the surprising observation that essential amino acid restriction results in the transcriptional derepression of silenced transgenes, including plasmid and oncoretroviral vectors in HeLa epithelial cells, and of HIV-1 provirus in T-lymphocytic cells. This effect correlates with a significant down-regulation of HDAC4. Consistently, HDAC4 pharmacological inhibition or knockdown reverts transcriptional silencing and leads to the reactivation of exogenous transgenes or of latent HIV-1. These results indicate that HDAC4 behaves as a critical regulator of exogenous transgene expression, sensitive to amino acid starvation, and suggest that selective HDAC4 inhibitors could represent unique and effective tools in the context of gene replacement, as well as of antiretroviral therapies.
Results
Amino Acid Starvation Induces Reversible Up-Regulation of Exogenous Transgene Expression.
This study stemmed from the previously reported finding that OA1 (GPR143, MIM 300808), a melanocyte-specific G protein-coupled receptor involved in ocular albinism (12), is sensitive to the presence of the essential amino acid Y in the culture medium (13). In particular, an OA1-GFP transgene was up-regulated in COS7 cells grown in the absence of Y or at low Y concentrations, suggesting that this amino acid could act similar to an agonist, leading to constitutive down-regulation of the receptor in regular culture medium (13). To establish the role of Y in OA1 function, we investigated the effects of Y deprivation in HeLa cells stably transfected with a plasmid vector (pcDNA3.1) expressing a mycHis-tagged human OA1 under the control of the CMV promoter (HeLa-OA1myc) by culturing them up to 5 d in medium deprived of either Y or other essential amino acids, such as methionine and cysteine (M/C), as a specificity control.
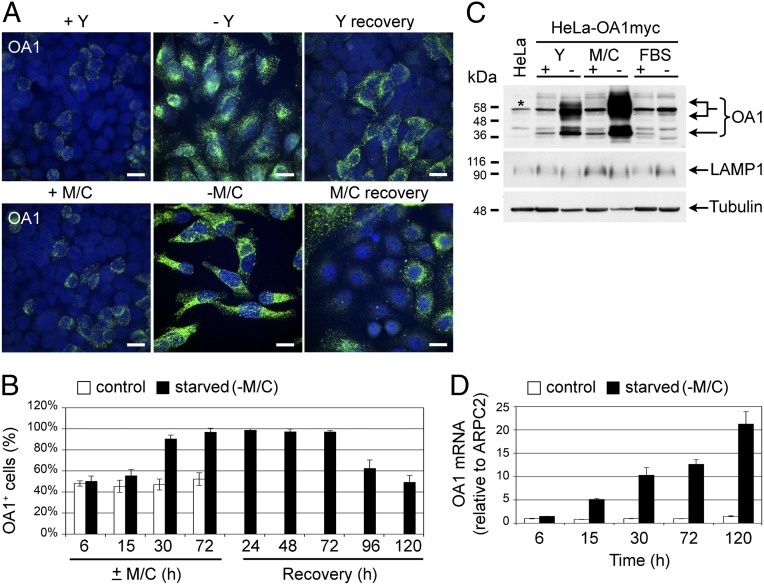
As often observed in transfected lines carrying integrated plasmids after long-term culture, about 50% of G418-resistant cells in the HeLa-OA1myc clonal population lost OA1 expression, as detectable by immunofluorescence (IF) analysis (Fig. 1A; +Y and +M/C). Surprisingly, not only Y but also M/C deprivation significantly up-regulated the expression of exogenous OA1, as demonstrated by both the brighter IF staining per cell and the higher percentage of expressing cells (Fig. 1A; −Y and −M/C). Indeed, on amino acid starvation, the number of OA1+ cells increased progressively, achieving nearly 100% in 3–5 d; in particular, M/C starvation induced a more rapid and marked up-regulation of OA1 than Y deprivation (Fig. 1B and Fig. S1A). This phenomenon was reversible, because when starved cells were recovered in complete medium, the brightness of OA1 staining declined considerably and the number of OA1+ cells returned to control conditions within 5 d (Fig. 1 A and B). Consistent with the IF results, the exogenous OA1 protein accumulated substantially on Y and especially M/C deprivation but not on serum deprivation (Fig. 1C).
Fig. 1.
OA1 transgene expression is reversibly up-regulated by amino acid starvation. (A) IF analysis of HeLa cells stably transfected with an expression vector for mycHis-tagged OA1 and clonally selected (HeLa-OA1myc). Cells were cultured in the presence (+) or absence (−) of Y or M/C for 5 d, followed by 5 d of recovery, and were then fixed and stained with anti-OA1 Ab (green) and Hoechst (blue). Both the intensity of the OA1 staining per cell and the number of OA1-expressing cells increase dramatically on starvation and decline on recovery. (Scale bars: 20 μm.) (B) Quantification of OA1-expressing cells in the HeLa-OA1myc population at the indicated times of M/C starvation and recovery. Results are expressed as a percentage of OA1+ cells of the total (mean ± SD of 5 random fields from 1 experiment representative of 4). (C) Immunoblotting of HeLa-OA1myc cell lysates after 5 d of culture in the presence (+) or absence (−) of Y, M/C, or FBS in the medium and decorated with specified Abs. Cell lysate from nontransfected HeLa cells not expressing OA1 serves as a negative control for the anti-OA1 Ab specificity (first lane). Note the huge accumulation of OA1 on amino acid but not FBS starvation compared with endogenous LAMP1 and tubulin. *Nonspecific product recognized by the anti-OA1 Ab. (D) Quantification of OA1 mRNA expression by real-time PCR in HeLa-OA1myc cells at the indicated times of M/C starvation. Data are expressed as the fold change compared with the amount of OA1 mRNA at 6 h in control conditions (mean ± SD of 3 technical replicates from 1 experiment representative of 3–4).
This effect was exclusively observed with exogenous proteins. In fact, in addition to OA1, other proteins exogenously expressed in HeLa cells under the control of the CMV promoter, such as rat LAMP1, displayed increased expression on either Y or M/C starvation (Fig. S1B). In contrast, the expression of endogenous LAMP1 in HeLa and melanoma cells and of endogenous OA1 in melanoma cells was unaffected or down-regulated on starvation (Fig. 1C and Fig. S1 B and C). This phenomenon was neither specific for integrated plasmids nor for proteins expressed under the control of the CMV promoter. Indeed, amino acid starvation elicited a similar effect in HeLa cells stably transduced with a retroviral vector expressing OA1 under the control of the Moloney murine leukemia virus LTR (HeLa-LOA1SN; Fig. S1D). Similarly, up-regulation of transgene expression on starvation was observed in HeLa clones stably transfected with plasmid vectors expressing OA1 under the control of the phosphoglycerate kinase 1 (PGK1) or elongation factor-1α (EF-1α) human promoters (HeLa-PGK1/OA1 and HeLa-EF1/OA1; Fig. S1 E and F).
Finally, the increased OA1 protein levels resulted from enhanced mRNA transcription and/or accumulation, because real-time PCR analysis of starved HeLa-OA1myc cells showed progressive increase of the OA1 mRNA levels up to over 20-fold vs. control after 5 d of M/C starvation (Fig. 1D). Thus, collectively, our results indicate that the up-regulation of OA1 on Y starvation originally reported by Lopez et al. (13) is neither specific for OA1 nor for Y deprivation. Rather, this phenomenon appears consequent to a general response of the cells to amino acid starvation, affecting the regulation of exogenous transgene expression under the control of either viral or human promoters through a mechanism that operates at the transcriptional/mRNA level.
Transgene Up-Regulation Is Induced by Histone Hyperacetylation but Not by DNA Hypomethylation.
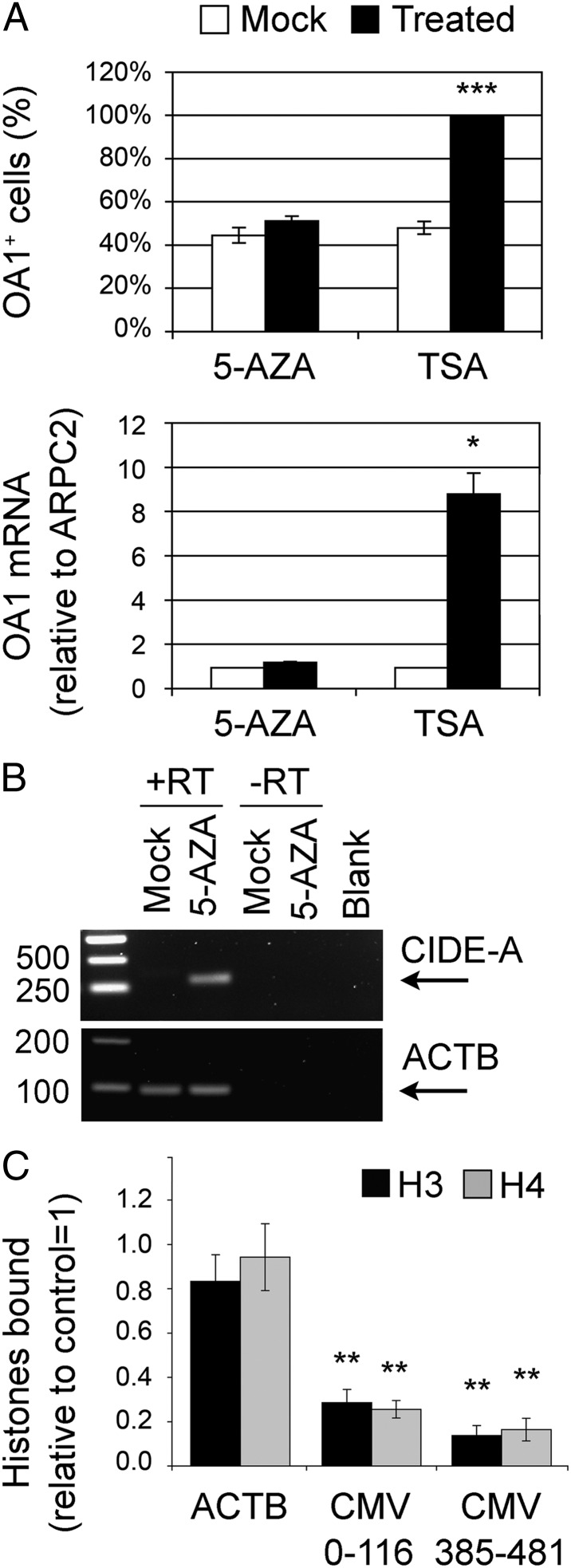
Amino acid deprivation strongly inhibits the activity of mammalian target of rapamycin (mTOR) kinase (14) and, if prolonged, causes cell growth delay and, eventually, death. Nevertheless, neither the inhibition of the mTOR pathway by incubation of HeLa-OA1myc cells with PP242 or rapamycin nor the activation of the cellular stress response by anisomycin or hydrogen peroxide increased the number of OA1+ cells (Fig. S2 A–F), suggesting that the up-regulated expression consequent to amino acid starvation depends on different mechanisms. Because long-term culture is frequently associated with epigenetic silencing of stably integrated exogenous transcriptional units, we tested if DNA methylation or histone deacetylation is involved in the regulation of transgene expression in our system. HeLa-OA1myc cells were incubated with pharmacological inhibitors of (i) DNA methylation, such as 5-aza-2′-deoxycytidine (5-AZA) (15), or (ii) HDACs, such as trichostatin A (TSA) (16). Inhibition of HDAC enzymatic activity by TSA for 15 h was sufficient to up-regulate the expression of exogenous OA1 to levels comparable to those obtained by 30 h of M/C starvation. In fact, OA1-expressing cells increased from 47% to almost 100%, and the OA1 mRNA levels were augmented about ninefold (Fig. 2A). Similar results were obtained by using sodium butyrate, another pan-HDACi (Fig. S2G). In contrast, incubation of the cells with 5-AZA for 72 h was completely ineffective (Fig. 2A), despite this treatment being sufficient to induce expression of the cell death-inducing DFF45-like effector A (CIDE-A) (17) (Fig. 2B).
Fig. 2.
Transgene up-regulation on starvation is reproduced by HDACi and is associated with histone displacement. (A) Assessment of OA1 expression in HeLa-OA1myc cells following treatment with TSA (pan-HDACi) for 15 h or with 5-AZA (DNA methylation inhibitor) for 72 h. (Upper) IF quantification of OA1-expressing cells, presented as a percentage of the total (mean ± SEM of 5 independent experiments; ***P < 0.001, paired two-tailed Student t test vs. mock). (Lower) Real-time PCR quantification of OA1 mRNA levels expressed as the fold change compared with the amount of OA1 mRNA in mock conditions (mean ± SEM of 3 independent experiments; *P < 0.05, paired two-tailed Student t test vs. mock). ARPC2, actin-related protein 2/3 complex, subunit 2. (B) PCR analysis of CIDE-A expression and ACTB in mock- and 5-AZA–treated cells to confirm treatment efficacy. Blank, amplification in the absence of template. (C) Quantification of histones H3 and H4 bound to the ACTB and CMV promoters after 30 h of M/C starvation relative to control conditions, as obtained by ChIP and real-time PCR analysis. Two pairs of primers were used for the CMV promoter, amplifying regions 0–116 and 385–481 (CMV total length = 654 bp). Data are normalized to the input DNA and expressed as the fold change vs. control (mean ± SEM of 3 independent experiments; **P < 0.01, paired two-tailed Student t test vs. control). The amount of H3 and H4 associated with both regions of the CMV promoter is significantly reduced after starvation.
To verify whether histone modifications and, in particular, acetylation at the CMV promoter were actually induced by amino acid deprivation, we performed ChIP experiments. Chromatin from control and starved (30 h without M/C) HeLa-OA1myc cells was immunoprecipitated with Abs to acetylated and total core histones H3 and H4, and the amount of coprecipitated CMV promoter was determined by real-time PCR using two pairs of primers covering different promoter regions. As a negative control, we also assessed the status of the endogenous β-actin (ACTB) promoter. Significantly, the amount of H3 and H4 bound to the CMV promoter decreased to about 15–30% on M/C starvation compared with control conditions (Fig. 2C). In contrast, the amount of histones H3 and H4 associated with the ACTB promoter and their levels in the total cell lysates remained similar in starved and control conditions (Fig. 2C and Fig. S2H). Moreover, a variable but specific increase in the acetylation state of both H3 and H4 on the CMV promoter but not ACTB promoter was observed (1.89 ± 0.37-fold and 3.53 ± 0.63-fold increase vs. control on the CMV promoter, compared with 0.90 ± 0.11-fold and 1.33 ± 0.16-fold on the ACTB promoter for acetylated H3 and H4, respectively; mean ± SEM of 3 independent experiments). Together, these data indicate that transgene up-regulation on amino acid deprivation is associated with epigenetic chromatin remodelling at the exogenous promoter, leading to transcriptional induction, and they point toward a role for HDACs in this process.
Amino Acid Starvation Determines a Specific Down-Regulation of HDAC4 Expression.
To search for the molecular players implicated in the cellular response to amino acid deprivation, we carried out a transcriptome analysis of nonclonal HeLa and clonal HeLa-OA1myc cell lines both in standard growth conditions and 5 d after either Y or M/C starvation. Of 48,803 transcript probes tested, 14,400 were ascertained to be expressed in HeLa cells based on a detection P value <0.01 recorded in at least one of the two conditions (i.e., control, starvation). Assuming that, except for OA1, both HeLa and HeLa-OA1myc cells would show a similar pattern of differentially expressed genes in response to amino acid limitation, we determined that of the 14,400 expressed transcripts, about 1,700 were significantly up-regulated, whereas 2,100 were significantly down-regulated in both cell types after starvation compared with controls (P < 0.05, paired Student t test). Furthermore, based on the transgene expression results, we expected that amino acid limitation would generate a stronger effect on M/C deprivation compared with Y deprivation. Thus, to narrow the number of possible candidates, we applied two types of fold-change/effect-size criteria by considering only (i) transcripts that were up- or down-regulated twofold or greater in starvation vs. control conditions and (ii) transcripts that showed a stronger (≥1.5-fold) effect in M/C vs. Y starvation.
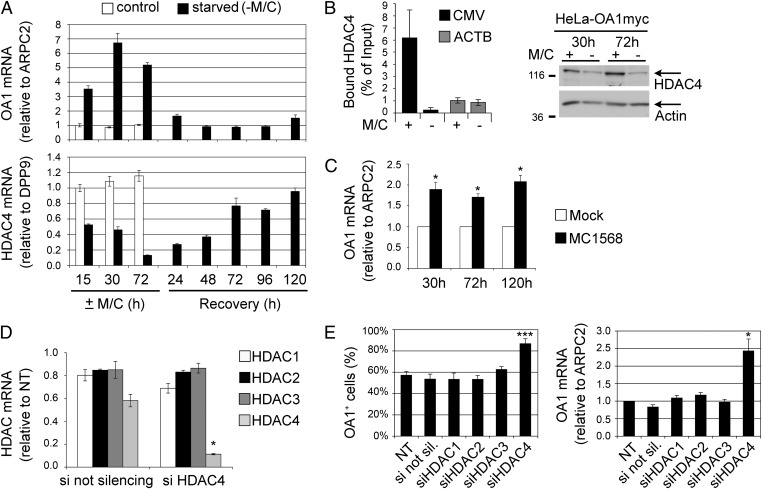
Based on these criteria, a total of 63 up-regulated and 216 down-regulated genes were selected and ranked according to their greater differential expression under M/C vs. Y deprivation. Interestingly enough, the second best candidate among down-regulated genes was a class II HDAC, namely HDAC4 (MIM 605314). Of note is the fact that among all expressed HDACs, HDAC4 was the only one that showed a substantial change in both amino acid starvation conditions and was also more modulated by M/C compared with Y deprivation. Indeed, HDAC4 expression was reduced by 50% in both HeLa and HeLa-OA1myc cells during Y deprivation and was completely suppressed by M/C starvation (Table 1). To validate these results, we performed a time course of M/C starvation and recovery in HeLa-OA1myc cells and analyzed the expression of OA1 and HDAC4 by real-time PCR. As shown in Fig. 3A, the expression of HDAC4 was progressively reduced to 13% of control levels after 72 h of starvation, concomitantly with the up-regulation of OA1. During recovery, HDAC4 expression increased progressively again, and it reached control levels after 72–120 h in normal complete medium, whereas OA1 expression switched off more rapidly, reaching levels comparable to those of control cells within 2 d. These findings reveal a strong inverse correlation between the expression of HDAC4 and that of exogenous OA1, suggesting the involvement of HDAC4 down-regulation in transgene derepression during amino acid starvation.
Table 1.
Differential expression of HDACs in starvation vs. control conditions
| Symbol | Av control* | Av starved* | t test† | Ctrl/Strv | +/−Y‡ | +/−M/C‡ | M/C vs. Y§ |
| HDAC1 | 12,497 ± 20% | 12,609 ± 27% | 0.97 | 0.99 | / | / | / |
| HDAC2 | 8,913 ± 9% | 9,190 ± 22% | 0.74 | 0.97 | / | / | / |
| HDAC3 | 2,425 ± 16% | 2,360 ± 12% | 0.68 | 1.03 | / | / | / |
| HDAC4 | 1,476 ± 26% | 341 ± 69% | 0.03 | 4.33 | 2.12 | 13.13 | 6.20 |
| HDAC6 | 587 ± 12% | 420 ± 5% | 0.01 | 1.40 | 1.39 | 1.41 | 1.01 |
| HDAC8 | 545 ± 7% | 502 ± 18% | 0.46 | 1.09 | / | / | / |
| SIRT1 | 1,516 ± 10% | 1,706 ± 34% | 0.49 | 0.89 | / | / | / |
| SIRT2 #1 | 254 ± 6% | 258 ± 9% | 0.80 | 0.98 | / | / | / |
| SIRT2 #2 | 301 ± 7% | 305 ± 10% | 0.85 | 0.98 | / | / | / |
| SIRT4 | 435 ± 30% | 931 ± 21% | 0.01 | 0.47 | 0.45 | 0.48 | 1.06 |
| SIRT5 #1 | 372 ± 11% | 281 ± 23% | 0.15 | 1.32 | / | / | / |
| SIRT5 #2 | 1,358 ± 9% | 666 ± 19% | 0.01 | 2.04 | 2.58 | 1.65 | 0.64 |
| SIRT7 | 470 ± 11% | 870 ± 33% | 0.09 | 0.54 | / | / | / |
Av, average; Ctrl, control; SIRT, sirtuin; Strv, starved; #, arbitrary number assigned to distinguish multiple probes referring to the same gene; /, not calculated.
*Average ± SD of signal values generated by Illumina microarrays for control or starved HeLa and HeLa-OA1myc cells.
†Paired two-tailed Student t test of starved vs. control group. Bold numbers indicate significant values and correspond to genes that are differentially expressed in the two conditions.
‡Ratio of mean signal values in control group (+) vs. starved group (−) for either Y or M/C deprivation conditions.
§Ratio of M/C vs. Y differential expression.
Fig. 3.
HDAC4 and OA1 expression correlate inversely during starvation, and HDAC4 inhibition or knockdown leads to OA1 transgene up-regulation. (A) Quantification of OA1 (Upper) and HDAC4 (Lower) mRNA expression by real-time PCR in HeLa-OA1myc cells at the indicated times of M/C starvation and recovery. Data are expressed as the fold change compared with the amount of the examined mRNA at 15 h in control conditions (mean ± SD of 3 technical replicates of 1 experiment representative of 3–4). Note the opposite trend of OA1 up-regulation and HDAC4 down-regulation. ARPC2, actin-related protein 2/3 complex, subunit 2. (B) Quantification of HDAC4 bound to the CMV (region 385–481) and ACTB promoters of HeLa-OA1myc cells after 30 h of M/C starvation relative to control conditions, as obtained by ChIP and real-time PCR analysis. Values were normalized to those obtained with rabbit IgG and expressed as a percentage of the input DNA (mean ± SEM of 2–3 independent experiments). HDAC4 is associated with the CMV promoter but not ACTB promoter in a starvation-sensitive manner. (Right) Immunoblotting analysis of HDAC4 protein levels after the indicated times of culture in the presence (+) or absence (−) of M/C. Actin was used as a loading control. (C) Quantification of OA1 mRNA expression by real-time PCR in HeLa-OA1myc cells at the indicated times of incubation with MC1568 (class II HDACi). Data are expressed as the fold change compared with the amount of the OA1 mRNA in mock conditions at each time point (mean ± SEM of 3 independent experiments; *P < 0.05, paired two-tailed Student t test vs. mock). (D) Quantification of HDAC1, HDAC2, HDAC3, and HDAC4 mRNA levels by real-time PCR following HDAC4 silencing (siHDAC4) compared with the not silencing siRNA (si not silencing). Data are expressed as the fold change compared with the amount of the examined mRNA in nontransfected cells (NT = 1; mean ± SEM of 3 independent experiments; *P < 0.05, paired two-tailed Student t test vs. the not silencing siRNA). (E) Assessment of OA1 expression in HeLa-OA1myc cells analyzed 72 h after transfection with the indicated siRNAs against different HDACs. (Left) IF quantification of OA1-expressing cells, presented as a percentage of the total (mean ± SEM of 4 independent experiments; ***P < 0.001, paired two-tailed Student t test vs. nontransfected cells). si not sil., siRNA not silencing. (Right) Real-time PCR quantification of OA1 mRNA levels expressed as the fold change compared with the amount of the OA1 mRNA in nontransfected cells (mean ± SEM of 5 independent experiments; *P < 0.05, paired two-tailed Student t test vs. nontransfected cells). Note that up-regulation of OA1 is obtained by HDAC4 silencing only.
HDAC4 Binds to the CMV Promoter, and Its Inhibition or Knockdown Leads to Increased Transgene Expression.
To assess whether HDAC4 was associated with the exogenous CMV promoter, we performed ChIP experiments in control conditions and after 30 h of M/C starvation. As shown in Fig. 3B, HDAC4 was detected at the CMV promoter but not at the ACTB promoter, and this binding decreased dramatically in starved conditions, even though the down-regulation of the enzyme induced by starvation was not yet complete at this time point. To investigate the functional role of HDAC4 in transgene derepression, we incubated HeLa-OA1myc cells with MC1568, a selective inhibitor of class II HDAC enzymatic activity (18). The treatment led to an increase of about twofold in the OA1 mRNA at every examined time point (Fig. 3C). This up-regulation was associated with an increased number of OA1+ cells from 50% to about 70%, and it was similarly reproduced by using MC1575, another class II HDACi (18) (Fig. S3A, Left). By contrast, pharmacological inhibition of HDAC6, representing the only other class II HDAC expressed in HeLa cells (Table 1), and of sirtuins, implicated in the longevity response to dietary restriction (19), failed to increase the number of OA1+ cells (Fig. S3A).
To confirm definitively the involvement of HDAC4, we specifically targeted its transcript by siRNAs. In parallel, we also silenced the abundant class I HDAC1, HDAC2, and HDAC3 as specificity controls. HeLa-OA1myc cells were transfected with a HDAC4 siRNA for 72 h and then analyzed by real-time PCR for HDAC transcript levels to assess siRNA specificity. As shown in Fig. 3D, the HDAC4 siRNA reduced the level of its target transcript to 11.4 ± 0.6% of the control untransfected cells, whereas it did not affect the expression of HDAC1, HDAC2, or HDAC3. Similarly, siRNA duplexes targeting HDAC1, HDAC2, and HDAC3 reduced the level of their targets to 10–15% compared with control untransfected cells (Fig. S3B). All siRNAs also induced significant down-regulation of the corresponding HDACs at the protein level (Fig. S3B). Next, we assessed the effect of siRNA-mediated HDAC silencing on OA1 expression. As shown in Fig. 3E, the down-regulation of HDAC4 but not that of HDAC1, HDAC2, and HDAC3 led to a significant increase in both the number of OA1+ cells within the HeLa-OA1myc cell population (from 57.2 ± 3.7% in untransfected cells to 86.8 ± 4.7% in HDAC4 siRNA-transfected cells) and the level of OA1 mRNA (2.5-fold). Similar results were obtained with another independent HDAC4 siRNA (Fig. S3 B and C). Together, these findings are consistent with a major role of HDAC4 in the reactivation process of epigenetically silenced transgenes triggered by amino acid starvation.
Amino Acid Starvation Reactivates Latent HIV-1 Provirus in HDAC4+ T-Lymphocytic ACH-2 but Not HDAC4− Promonocytic U1 Cells.
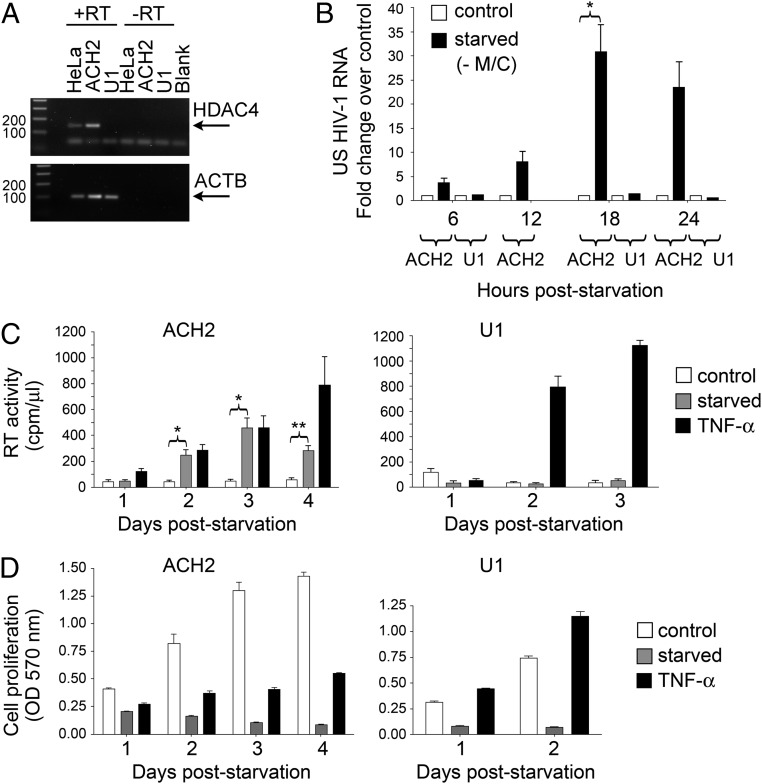
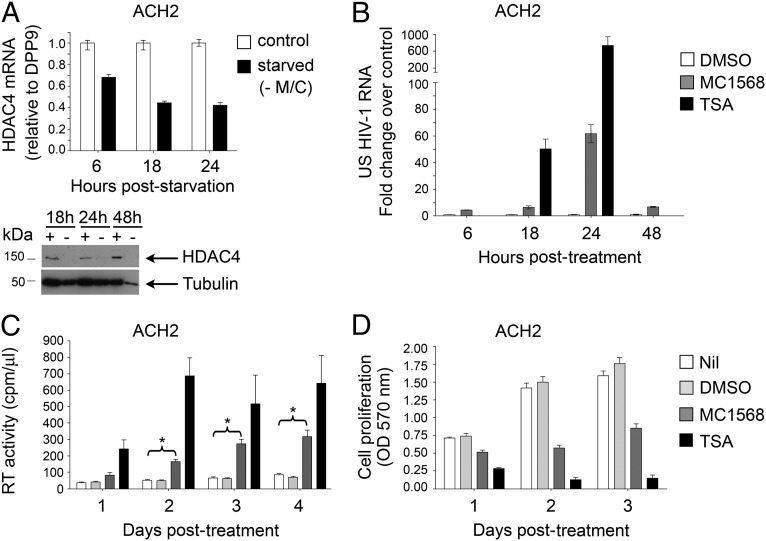
The role of epigenetic modifications in maintaining HIV-1 latent infection has been previously demonstrated (10). Indeed, broad-spectrum HDACi have been shown to reverse silencing and reactivate the dormant virus effectively, at least in tissue culture, and they are currently under investigation for clinical use (20, 21). Therefore, we investigated whether the pathway triggered by deprivation of essential amino acids, and particularly the inhibition of HDAC4, might be effective in promoting HIV-1 reactivation in infected cells. For this purpose, we first analyzed the expression level of HDAC4 mRNA in two well-characterized models of latent HIV-1 infection: the T-lymphocytic ACH-2 and promonocytic U1 cell lines, in which the almost undetectable virus expression can be significantly boosted by a variety of stimuli, including phorbol esters, cytokines, and broad-spectrum HDACi (22–26). As shown in Fig. 4A, HDAC4 was markedly expressed in ACH-2 cells, but it was undetectable in U1 cells, consistent with earlier studies in the U1-parental cell line U937 (27).
Fig. 4.
M/C starvation reactivates dormant HIV-1 provirus in HDAC4+ ACH-2 cells but not in HDAC4− U1 cells. (A) PCR amplification of HDAC4 and ACTB transcripts in HeLa, ACH-2, and U1 cells; HDAC4 is detected in ACH-2 cells but not in U1 cells. Blank, amplification in the absence of template. (B) Real-time PCR quantification of US HIV-1 RNA in ACH-2 and U1 cells cultivated in the presence (control) or absence of M/C for the indicated times. Results are expressed as the fold change vs. the amount of HIV-1 RNA in control conditions at each time point (mean ± SEM of 2–4 independent experiments; *P < 0.05, paired two-tailed Student t test vs. control cells). HIV-1 RNA expression was significantly up-regulated in starved ACH-2 cells but not in U1 cells. (C) RT activity of daily collected supernatants from ACH-2 and U1 cells cultivated up to 4 d in control medium in the absence of M/C (starved) or presence of TNF-α (mean ± SEM of 5 independent experiments for ACH-2 or 2 independent experiments for U1; *P < 0.05 and **P < 0.01, paired two-tailed Student t test vs. control cells). RT activity accumulation after M/C starvation was observed in ACH-2 cells only. (D) 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay performed on daily collected supernatants from ACH-2 or U1 cells cultured as described in C (mean ± SEM of 3 technical replicates of 1 experiment representative of 3 for ACH-2 or 2 for U1). M/C starvation affects viability and proliferation of ACH-2 and U1 cells to a comparable extent.
We then cultivated both ACH-2 and U1 cells in medium deprived of M/C, a condition that induced the most potent and rapid reactivation of repressed transgenes in HeLa cells. Consistent with the HDAC4 expression pattern, M/C deprivation strongly reactivated HIV-1 mRNA transcription in ACH-2 cells but not in U1 cells, reaching a peak of about 30-fold over control after 18 h of starvation (Fig. 4B). The increased level of transcription observed in ACH-2 cells was confirmed in terms of reverse transcriptase (RT) activity released in culture supernatant, which reached a peak 3 d after M/C starvation and was comparable to that induced by stimulation of the cells with tumor TNF-α (Fig. 4C, Left). In contrast, U1 cells responded well to TNF-α in terms of RT activity production but were unaffected by amino acid starvation (Fig. 4C, Right). The different responses of ACH-2 and U1 cells were not accounted for by a differential susceptibility to amino acid starvation-mediated cytostatic or cytotoxic effects in that both cell lines showed a similarly reduced cell proliferation in medium deprived of M/C (Fig. 4D). Overall, these observations are suggestive for a role of HDAC4 in HIV-1 reactivation on amino acid deprivation, similar to what was observed in HeLa cells.
HDAC4 Inhibition Leads to HIV-1 Transcriptional Activation and RT Activity Production in ACH-2 Cells.
We next investigated the levels of HDAC4 expression in ACH-2 cells under starvation or control conditions. As in HeLa cells, the expression of HDAC4 mRNA gradually decreased in the absence of M/C and its protein levels became almost undetectable (Fig. 5A), correlating inversely with the increased viral RNA accumulation (Fig. 4B). To test the involvement of HDAC4 in HIV-1 transcriptional reactivation directly, ACH-2 cells were incubated with the selective class II HDACi MC1568, and this resulted in a significant increase of HIV-1 mRNA at all tested time points, with a peak of 60-fold after 24 h of incubation (Fig. 5B), and a progressive accumulation of RT activity in the culture supernatants at up to 4 d of incubation (Fig. 5C). The kinetics of viral RNA accumulation induced by MC1568 were similar to those observed under starvation-dependent HDAC4 down-regulation (Figs. 4B and 5B, respectively), further supporting the hypothesis that these two pathways are strongly correlated. Moreover, incubation of ACH-2 cells with MC1568 produced an evident increase of the acetylation levels of histone H3 at the LTR region without affecting the total histone bound, similar to what was observed with the pan-HDACi TSA (Fig. S4A). Overall, TSA showed a more potent effect than MC1568 in terms of induction of both HIV-1 mRNA transcription and RT activity production (Fig. 5 B and C); however, it also caused an earlier and more dramatic loss of cell viability (Fig. 5D).
Fig. 5.
HDAC4 is down-regulated during starvation, and its inhibition leads to HIV-1 reactivation in ACH-2 cells. (A) Real-time PCR quantification of HDAC4 mRNA expression in ACH-2 cells in control medium or after M/C deprivation (starved) at the indicated times. The results are expressed as the fold change vs. control at each time point (mean ± SEM of 3 technical replicates of 1 experiment representative of 3). (Lower) Immunoblotting analysis of HDAC4 protein levels on whole-cell lysates extracted from ACH-2 cells cultivated in the presence (+) or absence (−) of M/C at the indicated times. Antitubulin Ab was used as a loading control. (B) Real-time PCR quantification of US HIV-1 RNA in ACH-2 cells incubated with DMSO (mock of MC1568), MC1568, or TSA at the reported times. Results are expressed as the fold change vs. the amount of US HIV-1 RNA in DMSO conditions at each time point (mean ± SEM of 3 technical replicates of 1 experiment representative of 3). MC1568, as well as the more potent TSA induced HIV-1 RNA transcription. (C) RT activity of daily collected supernatants from ACH-2 cells incubated with standard medium (Nil), DMSO, MC1568, or TSA for 4 d (mean ± SEM of 3 independent experiments; *P < 0.05, paired two-tailed Student t test vs. control). (D) 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay for cell viability and proliferation of daily collected supernatants from ACH-2 cells incubated for 3 d as described in C (mean ± SEM of 3 independent experiments).
Results similar to those obtained with MC1568 were observed with MC1575, a different class II HDACi, in HDAC4+ ACH-2 cells (Fig. S4 B and C), whereas no effects were achieved with MC1568 in HDAC4− U1 cells (Fig. S4D) or with HDAC6 inhibitors MC2726 or MC2727 in ACH-2 cells (Fig. S4 E–G). Altogether, these observations indicate that either M/C starvation or class II HDAC inhibition reactivates HIV-1 expression in ACH-2 cells but not in U1 cells via a mechanism not dependent on cytotoxic effects but possibly mediated by histone acetylation at the LTR promoter resulting from HDAC4 inactivation.
Discussion
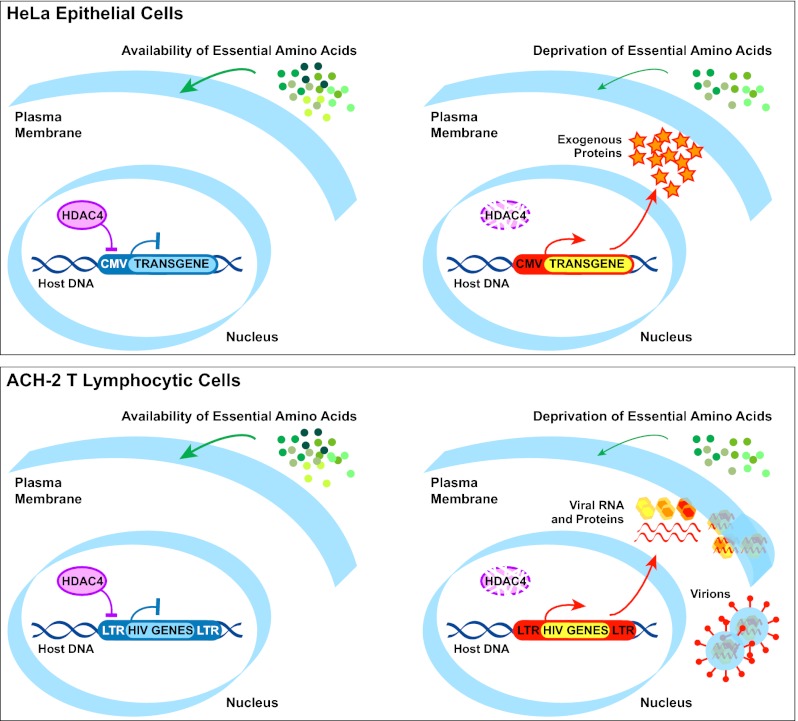
The previously reported finding that OA1 expression was up-regulated in the absence of Y in the culture medium was instrumental to support the notion that l-dopa is the endogenous ligand for this receptor, because Y could act by structurally mimicking l-dopa, leading to constitutive internalization and degradation of OA1 in standard growth conditions (13). At variance with this interpretation, we found that not only the absence of Y but the limitation of other essential amino acids, such as M/C, induces the up-regulation of OA1 and other exogenous transgenes. These can be expressed by means of viral or human promoters, and they can be carried by plasmid or retroviral vectors, which integrate into the genome by different mechanisms, typically generating multiple repeats or single proviral copies per site, respectively (28). In contrast, none of the corresponding endogenous proteins, including OA1, was up-regulated. These results indicate that amino acid starvation does not play a specific role in the function of the OA1 receptor; rather, it triggers a general epigenetic response able to derepress integrated transgenes, which could possibly be recognized by the presence of specific alterations in the chromatin structure established during insertion and silencing and then inherited.
The epigenetic silencing of exogenous sequences integrated into the genome represents a defensive mechanism of mammalian cells against retroviruses and other genetic parasitic elements. However, this physiological reaction can also repress transcription of oncoretroviral and lentiviral vectors carrying therapeutic transgenes, influencing the success of gene therapy approaches for the cure of inherited diseases (29). Furthermore, pathogenic retroviruses, such as HIV-1, can acquire a dormant state as integrated proviral DNA in a subset of infected cells to escape immunological or pharmacological surveillance (30). Therefore, the identification of mechanisms responsible for the reactivation of silenced transgenes bears potential clinical relevance. Among putative candidate pathways possibly involved in starvation-induced transgene up-regulation, we found that HDAC pan-inhibitors closely reproduced the starvation effects, suggesting that HDAC-mediated chromatin remodelling at the exogenous promoter might result in transcriptional induction.
By microarray expression analysis, we identified HDAC4 as a potential player in this process and time course analysis of OA1 and HDAC4 expression during M/C starvation and recovery showed a strong inverse correlation between the two transcripts, which displayed a similar dynamic and reversible yet opposite behavior. The involvement of HDAC4 was demonstrated by its association with the CMV promoter and, most importantly, by the use of pharmacological inhibitors and siRNA-mediated silencing, indicating that both inhibition and knockdown of HDAC4 are sufficient to reactivate transgene expression. The different kinetics of OA1 and HDAC4 expression during cell recovery after starvation and the reduced effect of HDAC4 inhibition or knockdown on exogenous OA1 up-regulation compared with amino acid deprivation suggest that this enzyme might not be the only player in the epigenetic response to starvation. Nevertheless, HDAC4 was the only HDAC expressed in HeLa cells whose specific down-regulation generated a significant up-regulation of exogenous OA1, supporting the notion that HDAC4 loss-of-function is critically implicated in mediating the reactivation of epigenetically silenced transgenes following amino acid limitation.
The mechanism by which HDAC4 regulates transgene silencing remains unclear. HDAC4 is a class IIa HDAC, highly expressed in brain, chondrocytes, heart, and skeletal muscle and playing a central role in bone development and myofiber gene expression (31, 32). It is generally accepted that class IIa HDACs lack significant HDAC activity per se, and may control gene transcription either by acting as scaffolds for complexes containing catalytically active class I HDACs (5) or by interacting with nucleoporins, thereby modifying chromatin association with the nuclear membrane (33). However, down-regulation of single class I HDACs did not reproduce HDAC4 inactivation, and the results obtained with enzymatic inhibitors of class II HDACs suggest that HDAC4 might maintain transgene silencing by means of its own catalytic activity. On the other hand, an alternative, and perhaps more likely, possibility is that the pharmacological inhibition of HDAC4 actually results in the displacement of interactors either by direct competition with proteins docked to the catalytic site by means of acetylated lysines, or by inducing a conformational change of the enzyme.
HIV-1 is a human pathogenic lentivirus infecting CD4+ T cells and mononuclear phagocytes, and causing AIDS (34, 35). Enormous advances have been made in the development of anti-HIV agents administered in protocols of cART; however, these can only control the capacity of HIV-1 to propagate to new target cells but cannot eradicate the reservoirs of chronically infected cells carrying integrated proviruses (36). Consequently, cART suspension almost invariably triggers the resurgence of viral replication and immunodeficiency (30). For these reasons, the search for strategies capable of affecting the pool of persistently infected cells, including those carrying latent proviruses, is among the highest priorities in the international research agenda (37). In this scenario, HDACi are intensely investigated in individuals receiving cART with the goal of inducing viral expression from latently infected cells so as to render them sensitive to either immunological or pharmacological purging strategies (9, 11, 38).
Therefore, we explored the possibility that amino acid starvation and HDAC4 inhibition could reactivate HIV-1 expression from latently infected cells. For this purpose, we selected two well-validated cellular models of HIV-1 postintegration latency, namely, ACH-2 and U1, of T-lymphocytic and promonocytic origin, respectively, to reflect the two main cellular reservoirs of HIV-1 in cART-treated individuals. Indeed, these cell lines are widely used in preclinical studies for new antilatency molecules capable of derepressing the proviral LTR and of being combined with cART (25, 39–41). However, we observed that although ACH-2 cells express HDAC4 constitutively, U1 cells do not. Consistent with a role for the enzyme, both the use of medium deprived from essential amino acids and pharmacological inhibition of class II HDACs reactivated HIV-1 expression and viral production in ACH-2 cells but not in U1 cells.
Although a role for class II HDACs has been previously proposed in maintaining the latency of episomal γ-herpesviruses (42), to the best of our knowledge, our findings suggest an unprecedented involvement of this class of deacetylases, and particularly of HDAC4, in the regulation of HIV-1 provirus latency and expression. Conversely, the importance of class I HDACs in repressing HIV-1 expression is highlighted by several studies demonstrating that viral outgrowth can be induced both from cell line models of latency and from resting CD4+ T cells of aviremic cART-treated patients, either by class I HDACi, such as valproic acid, or by pan-inhibitors, such as suberoylanilide hydroxamic acid (Vorinostat) (20, 21, 25). Nevertheless, the clinical use of broad-spectrum HDACi has been limited so far by their toxicity, leading to relevant side effects (6). By contrast, the class II HDACi tested here induced a significant up-regulation of HIV-1 expression coupled with limited cytotoxicity, even at relatively high concentrations. Therefore, although future studies will be needed to evaluate the impact of HDAC4 targeting on patients’ viral reservoirs, likely in combination with other antilatency strategies to overcome response heterogeneity (30, 40), our findings provide a proof of concept that specific HDAC4 inhibitors bear potential relevance as effective and safer antilatency weapons.
In conclusion, our study unveils an unsuspected link between a selective dietary restriction and the epigenetic regulation of transgene expression, provides insights into the regulatory mechanisms that supervise the silencing of potentially dangerous exogenous sequences integrated into the genome, and suggest that HDAC4 might represent a unique therapeutic target in the context of gene replacement as well as antiretroviral strategies. Finally, our findings suggest that malnutrition, and particularly protein deficiency, might contribute to promoting HIV-1 proviral reactivation and spreading, a poorly explored area that deserves future investigation.
Materials and Methods
Cell Lines and Media.
HeLa epithelial and M14 melanoma cells were maintained in DMEM, and MEL-SK28 melanoma cells were maintained in RPMI 1640, both containing glutaMAX (Invitrogen) and supplemented with 10% (vol/vol) FBS (Sigma), 100 U/mL penicillin G (Invitrogen), and 100 μg/mL streptomycin (Invitrogen), at 37 °C in a 5% (vol/vol) CO2 humidified atmosphere. The ACH-2 and U1 cell lines (22, 23) carry one and two copies of integrated proviruses, respectively, characterized by a state of relative latency, and were cultured as described for U1 (26). All cell lines are of human origin.
Reagents and Cell Treatments.
PP242, anisomycin, hydrogen peroxide, TSA, sodium butyrate, 5-AZA, and nicotinamide (NAM) were from Sigma; rapamycin was from Cell Signaling; MC1568 (18) was from Selleck Chemicals; and MC1575 (18), MC2726 (compound 7), and MC2727 (compound 3) (43) were from the laboratory of one of the authors (A.M.). Recombinant TNF-α was from R&D Systems. Drugs were used at the following final concentrations: PP242, rapamycin, and TSA at 1 μM; anisomycin at 125 nM; hydrogen peroxide at 50 μM; 5-AZA at 2 μM; sodium butyrate at 1 mM; NAM at 20 mM; MC1568 at 20 μM in HeLa cells and 5–10 μM in ACH-2 cells (with similar results); MC1575 at 20 μM in HeLa cells and 5 μM in ACH-2 cells; and MC2726 and MC2727 at 0.1 μM. TNF-α at 1 ng/mL; vehicle was used as a mock control.
Amino Acid Starvation.
DMEM deprived of M/C, RPMI deprived of Y, and dialyzed FBS were from Invitrogen. HeLa, M14, and SK-MEL28 cells were seeded at 30–40% confluency; cells to be starved for more than 30 h were plated more confluently with respect to the control. The following day, cells were washed and cultured in the appropriate medium, with or without M/C or Y, for 6 h up to 5 d. Starting from the second or third day of starvation, dead cells were removed by gentle aspiration and fresh medium was added. ACH-2 or U1 cells were washed twice with PBS and then resuspended in the appropriate medium, with or without M/C. The cells were seeded at 200,000 cells/mL and cultured from 6 to 96 h.
Expression Vectors, Stable Transfection, and Retroviral Infection.
pcDNA3.1/OA1myc-His, pCR3/ratLAMP1, and LOA1SN were previously described (44, 45). The OA1 expression plasmids pPGK1/OA1 and pEF1/OA1 were generated by inserting the PGK1 and EF-1α human promoters in the pCR3 and pcDNA3.1 vectors (Invitrogen), respectively, in place of the CMV promoter, which was excised away. The PGK1 and EF-1α human promoters were a kind gift of D. Cesana and E. Montini (Telethon Institute for Gene Therapy, Milan, Italy). For stable transfection, HeLa cells were transfected using FuGENE 6 (Roche) and selected with 800 μg/mL G418 (Sigma), which was maintained thereafter to avoid loss of plasmid integration. Resistant clones were isolated and analyzed for protein expression by IF. For retroviral infection, the LOA1SN retrovirus was produced in Phoenix ampho packaging cells, following the protocol by Dr. Garry Nolan, at Stanford University. HeLa cells were infected as described (45) and selected as a polyclonal population.
siRNA Design and Transfection.
siRNA against HDACs was designed using an algorithm supplied at http://sirna.wi.mit.edu/ or were modified from Mottet et al. (46) (HDAC3 and HDAC4 #1). The sequences are provided in Table S1. Cells were transfected with 50 nM siRNA and 1 μL of Lipofectamine 2000 (Invitrogen), following the manufacturer’s instructions, at days 1 and 3 postplating. The medium was changed 24 h after each transfection, and the analysis was carried out 72 h after the first transfection.
IF.
IF was carried out as described (44, 45, 47). Cells were observed with a UApo\340 40× oil objective (Olympus; n.a. of 1.35) on an Olympus IX70 microscope (DeltaVision System; Applied Precision), and images were acquired using softWoRx 3.5 software (Applied Precision) and processed as previously described (45). The percentage of OA1-expressing cells was evaluated by IF analysis by counting the number of expressing cells out of the total number of cells in five random 63× fields.
Western Immunoblotting.
Western immunoblotting was carried out as described (12, 47). For histones and HDACs, cells were lysed in Laemmli buffer, boiled at 95 °C for 5 min, sonicated for 30 s at 100% power (Bandelin Sonopuls HD2200 water sonicator), boiled again for 5 min, and resolved on a 7.5–15% (wt/vol) polyacrylamide gel. ACH-2 cells were lysed, and proteins were separated as described (26). Primary Abs were as follows: anti-human OA1 (47); H4A3 anti-human Lamp1 and E7 anti–β-tubulin (Hibridoma Databank); anti-rat Lamp1 (gift of Yoshitaka Tanaka, Kyushu University, Fukuoka, Japan); AC-15 anti–β-actin (Sigma); 5G10 anti-S6 ribosomal protein and anti–phospho-S6 ribosomal protein (Ser240/244) (Cell Signaling); anti–phospho-p38 (Tyr180/182) (Invitrogen); anti-p38 and H92 anti-HDAC4 (Santa Cruz); anti-HDAC2 and anti-HDAC3 (Abcam); and anti-HDAC1 (Millipore).
Microarray Analysis.
HeLa and HeLa-OA1myc cells were starved for 5 d in absence of Y or M/C. Total RNA was extracted with the RNeasy minikit (Qiagen). The gene expression profile was determined through Illumina BeadArray technology, following the manufacturer’s instructions and using the Illumina TotalPrep RNA Amplification Kit (Applied Biosystems), the Illumina HumanWG-6 BeadChips v.3 (48,803 transcripts), and the Illumina BeadArray Reader. The software Illumina BeadStudio v.3 was used to assess the system quality controls and to elaborate the fluorescence signal to an intensity value corresponding to the transcript amount. Gene expression data were normalized using the quantile algorithm in the software, and only transcripts whose intensity value was significantly different from that of background (detection P value <0.01) in at least one sample have been considered in the statistical analyses. Data have been submitted to Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo, accession no. GSE35323).
cDNA Synthesis and Real-Time PCR.
An equal amount (1 μg) of RNA from HeLa cells was reverse-transcribed using the SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen) with oligo-dT as primers and diluted to 5 ng/μL. The cDNA (3 μL) was amplified by real-time PCR using SYBR Green Master Mix on a Light Cycler 480 (Roche) according to the manufacturer’s instructions. The thermal cycling conditions were 1 cycle at 95 °C for 5 min, followed by 40–45 cycles at 95 °C for 20 s, 56 °C or 54 °C for 20 s, and 72 °C for 20 s. The sequences, efficiencies, and annealing temperatures of the primers are provided in Table S2. Data were analyzed with Microsoft Excel using the 2-ΔΔct method (48) or the formula EtargetΔct target (control − sample)/EreferenceΔct reference (control − sample) in the case of OA1 quantification (49). Reference genes for normalization, showing no major changes in starvation vs. control conditions by microarray analysis, were actin-related protein 2/3 complex, subunit 2 for highly expressed transcripts (OA1 and HDAC1-2) and dipeptidyl-peptidase 9 for poorly expressed transcripts (HDAC3 and HDAC4).
Conventional PCR Amplification.
PCR conditions for HDAC4 and HDAC6 were as follow: 1 cycle at 95 °C for 5 min, followed by 40 cycles at 95 °C for 20 s, 56 °C for 20 s, and 72 °C for 20 s, and a final extension step of 10 min at 72 °C. PCR conditions for ACTB were as follows: 1 cycle at 95 °C for 5 min, followed by 30 cycles at 95 °C for 20 s, 56 °C for 20 s, and 72 °C for 20 s, and a final extension step of 10 min at 72 °C. PCR conditions for cell-death-inducing DFF45-like effector A (17) were as follow: 1 cycle at 95 °C for 5 min, followed by 35 cycles at 95 °C for 60 s, 58 °C for 60 s, and 72 °C for 90 s, and a final extension step of 10 min at 72 °C. The primer sequences are provided in Table S2.
ChIP.
HeLa-OA1myc cells were starved in M/C-free medium for 30 h. ACH-2 cells were incubated with either TSA or MC1568 for 18 h and 24 h, respectively. Chromatin preparation and ChIP were performed according to the method of Lee et al. (50), using 10 μL of anti-H4 (catalog no. 05-858; Millipore) or anti–acetyl-H4 (catalog no. 06-866; Millipore), 5 μg of anti-H3 (catalog no. ab1791; Abcam) or anti–acetyl-H3 (catalog no. 06-599; Millipore), or 10 μg of H92 anti-HDAC4 (Santa Cruz) or rabbit IgG (catalog no. 011-000-003; Jackson ImmunoResearch) overnight at 4 °C. After elution and cross-linking reversal, DNA was purified using the Qiaquik PCR purification kit (Qiagen). HeLa-OA1myc DNA was analyzed by real-time PCR using two pairs of primers for the CMV promoter (amplifying regions 1–116 and 385–481) and one pair for the ACTB promoter (Table S2). Genomic DNA from untransfected HeLa was used as a negative control. ACH-2 DNA was amplified by real-time PCR using PCR1 and PCR2 (51) primers specific for the LTR promoter and, as a negative control, B13 primers specific for a genomic region not containing binding sites for known transcription factors (52). Values were normalized to their input control by using the 2-Δct equation (48), and histone acetylation levels were calculated as the ratio between acetylated vs. total histones. HDAC4 values were normalized to those obtained from rabbit IgG and expressed as a percentage of the input.
Quantification of HIV-1 RNA Transcripts.
Total RNA was extracted from ACH-2 and U1 cells by TRIZOL and the Pure link Micro-to-Midi Total RNA Purification System (Invitrogen). The RNA (1 μg) was digested with DNase I following the manufacturer’s instructions (Roche). cDNA synthesis was carried out using the SuperScript First-Strand Synthesis System for RT-PCR as previously described (52), with the exception that SuperScript II RT (50 units) was used. Unspliced (US) HIV-1 RNA, including both genomic and mRNA, was quantified by TaqMan assay with a 7500 Fast Real-Time PCR system (Applied Biosystems). The cDNA (50 ng) was amplified by real-time PCR using primers and a probe that recognize the HIV-1 gag gene, as described by Vicenzi et al. (53) (Table S2). The 18S and GAPDH transcripts were amplified with a commercial kit following the manufacturer’s procedures (Applied Biosystems) and were used for normalization. The thermal cycling conditions were 50 °C for 2 min, 95 °C for 15 min, and 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The levels of the US HIV-1 RNA were calculated by the 2-ΔΔct method (48).
RT Activity Assay.
HIV production was measured in cell culture supernatants by a liquid-phase Mg2+-dependent RT activity assay, as described previously (53).
3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide Proliferation and Viability Assay.
Cells were seeded in 96-well plates at 4 × 104 cells per well (in 200 μL per well); after treatments, cell growth and viability were tested by the [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay according to the manufacturer’s instructions (Roche). The absorbance within the cells was quantified at 570 nm (background correction at 655 nm) using an ELISA reader (Bio-Rad). The data are presented as ODs.
Statistical Analyses.
All calculations and statistical analyses were carried out using Microsoft Excel or GraphPad Prism software.
Supplementary Material
Acknowledgments
G.D.C. performed the vast majority of HIV-related experiments as partial fulfillment of her doctoral degree in Basic and Applied Immunology, Vita-Salute San Raffaele University, School of Medicine, Milan. We thank several colleagues within and outside our institution for helpful discussions and suggestions. We also thank Dr. S. Ghezzi for technical help. Part of this work was carried out in Alembic, Imaging Facility of Ospedale San Raffaele and Vita-Salute University, Milan. This work was supported by Telethon-Italy Grant GGP08156 (to M.V.S) and Grant TCP05001 (to D.G.); the Vision of Children Foundation (San Diego, CA) (to M.V.S.); Cassa di Risparmio delle Provincie Lombarde Foundation Grant 2008-2230 (to G.P.); Italian Ministry of Health Grant Program of AIDS Research 2009-2010 (to G.P.) and Grant GRO8-21 (to D.G.); Progetto Giovani Ricercatori 2007 D.lgs 502/92 (to F.M.B.); European Research Council Grant StG 204279 (to D.G.); Muscular Dystrophy Association Grant MDA115400 (to D.G.); the Facioscapulohumeral Muscular Dystrophy Society (to D.G.); Italian Ministry of University and Research Grant PRIN 2009PX2T2E (to A.M.); and European Commission Seventh Framework Programme Grant BLUEPRINT/282510 (to A.M.).
Footnotes
The authors declare no conflict of interest.
This Direct Submission article had a prearranged editor.
Data deposition: The microarray data reported in this paper have been deposited in to the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE35323).
See Author Summary on page 13482 (volume 109, number 34).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1202174109/-/DCSupplemental.
References
- 1.Kilberg MS, Pan YX, Chen H, Leung-Pineda V. Nutritional control of gene expression: How mammalian cells respond to amino acid limitation. Annu Rev Nutr. 2005;25:59–85. doi: 10.1146/annurev.nutr.24.012003.132145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet. 2010;11:285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lahm A, et al. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc Natl Acad Sci USA. 2007;104:17335–17340. doi: 10.1073/pnas.0706487104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischle W, et al. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- 6.Prince HM, Bishton MJ, Harrison SJ. Clinical studies of histone deacetylase inhibitors. Clin Cancer Res. 2009;15:3958–3969. doi: 10.1158/1078-0432.CCR-08-2785. [DOI] [PubMed] [Google Scholar]
- 7.Mai A, Rotili D, Valente S, Kazantsev AG. Histone deacetylase inhibitors and neurodegenerative disorders: Holding the promise. Curr Pharm Des. 2009;15:3940–3957. doi: 10.2174/138161209789649349. [DOI] [PubMed] [Google Scholar]
- 8.Colussi C, et al. Histone deacetylase inhibitors: Keeping momentum for neuromuscular and cardiovascular diseases treatment. Pharmacol Res. 2010;62:3–10. doi: 10.1016/j.phrs.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 9.Lewin SR, Rouzioux C. HIV cure and eradication: How will we get from the laboratory to effective clinical trials? AIDS. 2011;25:885–897. doi: 10.1097/QAD.0b013e3283467041. [DOI] [PubMed] [Google Scholar]
- 10.Hakre S, Chavez L, Shirakawa K, Verdin E. Epigenetic regulation of HIV latency. Curr Opin HIV AIDS. 2011;6:19–24. doi: 10.1097/COH.0b013e3283412384. [DOI] [PubMed] [Google Scholar]
- 11.Margolis DM. Histone deacetylase inhibitors and HIV latency. Curr Opin HIV AIDS. 2011;6:25–29. doi: 10.1097/COH.0b013e328341242d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schiaffino MV, et al. Ocular albinism: Evidence for a defect in an intracellular signal transduction system. Nat Genet. 1999;23:108–112. doi: 10.1038/12715. [DOI] [PubMed] [Google Scholar]
- 13.Lopez VM, Decatur CL, Stamer WD, Lynch RM, McKay BS. L-DOPA is an endogenous ligand for OA1. PLoS Biol. 2008;6:e236. doi: 10.1371/journal.pbio.0060236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patra SK, Patra A, Rizzi F, Ghosh TC, Bettuzzi S. Demethylation of (Cytosine-5-C-methyl) DNA and regulation of transcription in the epigenetic pathways of cancer development. Cancer Metastasis Rev. 2008;27:315–334. doi: 10.1007/s10555-008-9118-y. [DOI] [PubMed] [Google Scholar]
- 16.Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 17.Li D, Da L, Tang H, Li T, Zhao M. CpG methylation plays a vital role in determining tissue- and cell-specific expression of the human cell-death-inducing DFF45-like effector A gene through the regulation of Sp1/Sp3 binding. Nucleic Acids Res. 2008;36:330–341. doi: 10.1093/nar/gkm1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mai A, et al. Class II (IIa)-selective histone deacetylase inhibitors. 1. Synthesis and biological evaluation of novel (aryloxopropenyl)pyrrolyl hydroxyamides. J Med Chem. 2005;48:3344–3353. doi: 10.1021/jm049002a. [DOI] [PubMed] [Google Scholar]
- 19.Lu SP, Lin SJ. Regulation of yeast sirtuins by NAD(+) metabolism and calorie restriction. Biochim Biophys Acta. 2010;1804:1567–1575. doi: 10.1016/j.bbapap.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lehrman G, et al. Depletion of latent HIV-1 infection in vivo: A proof-of-concept study. Lancet. 2005;366:549–555. doi: 10.1016/S0140-6736(05)67098-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Archin NM, et al. Expression of latent HIV induced by the potent HDAC inhibitor suberoylanilide hydroxamic acid. AIDS Res Hum Retroviruses. 2009;25:207–212. doi: 10.1089/aid.2008.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Folks TM, et al. Characterization of a promonocyte clone chronically infected with HIV and inducible by 13-phorbol-12-myristate acetate. J Immunol. 1988;140:1117–1122. [PubMed] [Google Scholar]
- 23.Clouse KA, et al. Monokine regulation of human immunodeficiency virus-1 expression in a chronically infected human T cell clone. J Immunol. 1989;142:431–438. [PubMed] [Google Scholar]
- 24.Poli G, Orenstein JM, Kinter A, Folks TM, Fauci AS. Interferon-alpha but not AZT suppresses HIV expression in chronically infected cell lines. Science. 1989;244:575–577. doi: 10.1126/science.2470148. [DOI] [PubMed] [Google Scholar]
- 25.Savarino A, et al. “Shock and kill” effects of class I-selective histone deacetylase inhibitors in combination with the glutathione synthesis inhibitor buthionine sulfoximine in cell line models for HIV-1 quiescence. Retrovirology. 2009;6:52. doi: 10.1186/1742-4690-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Della Chiara G, et al. Negative regulation of HIV-1 transcription by a heterodimeric NF-κB1/p50 and C-terminally truncated STAT5 complex. J Mol Biol. 2011;410:933–943. doi: 10.1016/j.jmb.2011.03.044. [DOI] [PubMed] [Google Scholar]
- 27.Inoue S, Mai A, Dyer MJ, Cohen GM. Inhibition of histone deacetylase class I but not class II is critical for the sensitization of leukemic cells to tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis. Cancer Res. 2006;66:6785–6792. doi: 10.1158/0008-5472.CAN-05-4563. [DOI] [PubMed] [Google Scholar]
- 28.Brown PO. Integration. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 29.Toscano MG, et al. Physiological and tissue-specific vectors for treatment of inherited diseases. Gene Ther. 2011;18:117–127. doi: 10.1038/gt.2010.138. [DOI] [PubMed] [Google Scholar]
- 30.Trono D, et al. HIV persistence and the prospect of long-term drug-free remissions for HIV-infected individuals. Science. 2010;329:174–180. doi: 10.1126/science.1191047. [DOI] [PubMed] [Google Scholar]
- 31.Vega RB, et al. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119:555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 32.Cohen TJ, et al. The histone deacetylase HDAC4 connects neural activity to muscle transcriptional reprogramming. J Biol Chem. 2007;282:33752–33759. doi: 10.1074/jbc.M706268200. [DOI] [PubMed] [Google Scholar]
- 33.Kehat I, Accornero F, Aronow BJ, Molkentin JD. Modulation of chromatin position and gene expression by HDAC4 interaction with nucleoporins. J Cell Biol. 2011;193:21–29. doi: 10.1083/jcb.201101046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barré-Sinoussi F, et al. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 35.Popovic M, Sarngadharan MG, Read E, Gallo RC. Detection, isolation, and continuous production of cytopathic retroviruses (HTLV-III) from patients with AIDS and pre-AIDS. Science. 1984;224:497–500. doi: 10.1126/science.6200935. [DOI] [PubMed] [Google Scholar]
- 36.Finzi D, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 37.Richman DD, et al. The challenge of finding a cure for HIV infection. Science. 2009;323:1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- 38.Demonté D, Quivy V, Colette Y, Van Lint C. Administration of HDAC inhibitors to reactivate HIV-1 expression in latent cellular reservoirs: Implications for the development of therapeutic strategies. Biochem Pharmacol. 2004;68:1231–1238. doi: 10.1016/j.bcp.2004.05.040. [DOI] [PubMed] [Google Scholar]
- 39.Choi BS, et al. Novel histone deacetylase inhibitors CG05 and CG06 effectively reactivate latently infected HIV-1. AIDS. 2010;24:609–611. doi: 10.1097/QAD.0b013e328333bfa1. [DOI] [PubMed] [Google Scholar]
- 40.Reuse S, et al. Synergistic activation of HIV-1 expression by deacetylase inhibitors and prostratin: Implications for treatment of latent infection. PLoS ONE. 2009;4:e6093. doi: 10.1371/journal.pone.0006093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fernandez G, Zeichner SL. Cell line-dependent variability in HIV activation employing DNMT inhibitors. Virol J. 2010;7:266. doi: 10.1186/1743-422X-7-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goodwin MM, et al. Histone deacetylases and the nuclear receptor corepressor regulate lytic-latent switch gene 50 in murine gammaherpesvirus 68-infected macrophages. J Virol. 2010;84:12039–12047. doi: 10.1128/JVI.00396-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kozikowski AP, Tapadar S, Luchini DN, Kim KH, Billadeau DD. Use of the nitrile oxide cycloaddition (NOC) reaction for molecular probe generation: A new class of enzyme selective histone deacetylase inhibitors (HDACIs) showing picomolar activity at HDAC6. J Med Chem. 2008;51:4370–4373. doi: 10.1021/jm8002894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piccirillo R, et al. An unconventional dileucine-based motif and a novel cytosolic motif are required for the lysosomal and melanosomal targeting of OA1. J Cell Sci. 2006;119:2003–2014. doi: 10.1242/jcs.02930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palmisano I, et al. The ocular albinism type 1 protein, an intracellular G protein-coupled receptor, regulates melanosome transport in pigment cells. Hum Mol Genet. 2008;17:3487–3501. doi: 10.1093/hmg/ddn241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mottet D, et al. HDAC4 represses p21(WAF1/Cip1) expression in human cancer cells through a Sp1-dependent, p53-independent mechanism. Oncogene. 2009;28:243–256. doi: 10.1038/onc.2008.371. [DOI] [PubMed] [Google Scholar]
- 47.Schiaffino MV, et al. The ocular albinism type 1 gene product is a membrane glycoprotein localized to melanosomes. Proc Natl Acad Sci USA. 1996;93:9055–9060. doi: 10.1073/pnas.93.17.9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 49.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marban C, et al. Recruitment of chromatin-modifying enzymes by CTIP2 promotes HIV-1 transcriptional silencing. EMBO J. 2007;26:412–423. doi: 10.1038/sj.emboj.7601516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lusic M, Marcello A, Cereseto A, Giacca M. Regulation of HIV-1 gene expression by histone acetylation and factor recruitment at the LTR promoter. EMBO J. 2003;22:6550–6561. doi: 10.1093/emboj/cdg631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vicenzi E, et al. Envelope-dependent restriction of human immunodeficiency virus type 1 spreading in CD4(+) T lymphocytes: R5 but not X4 viruses replicate in the absence of T-cell receptor restimulation. J Virol. 1999;73:7515–7523. doi: 10.1128/jvi.73.9.7515-7523.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]