Abstract
Vertebrate Hedgehog (Hh) signals involved in development and some forms of cancer, such as basal cell carcinoma, are transduced by the primary cilium, a microtubular projection found on many cells. A critical step in vertebrate Hh signal transduction is the regulated movement of Smoothened (Smo), a seven-transmembrane protein, to the primary cilium. To identify small molecules that interfere with either the ciliary localization of Smo or ciliogenesis, we undertook a high-throughput, microscopy-based screen for compounds that alter the ciliary localization of YFP-tagged Smo. This screen identified 10 compounds that inhibit Hh pathway activity. Nine of these Smo antagonists (SA1–9) bind Smo, and one (SA10) does not. We also identified two compounds that inhibit ciliary biogenesis, which block microtubule polymerization or alter centrosome composition. Differential labeling of cell surface and intracellular Smo pools indicates that SA1–7 and 10 specifically inhibit trafficking of intracellular Smo to cilia. In contrast, SA8 and 9 recruit endogenous Smo to the cilium in some cell types. Despite these different mechanisms of action, all of the SAs inhibit activation of the Hh pathway by an oncogenic form of Smo, and abrogate the proliferation of basal cell carcinoma-like cancer cells. The SA compounds may provide alternative means of inhibiting pathogenic Hh signaling, and our study reveals that different pools of Smo move into cilia through distinct mechanisms.
Keywords: high-throughput screening, high-content analysis, fluorescence cell-based assay, Sufu, medulloblastoma
Hedgehog (Hh) glycolipoproteins signal through a pathway partially conserved from Drosophila to humans and crucial for regulating tissue patterning and cell cycle regulation. In adult tissues, inappropriate activation of the Hh pathway is associated with medulloblastoma and basal cell carcinoma (BCC) (1–4). Similarly, inheritance of a loss-of-function allele of Patched1 (Ptch1), a negative regulator of Hh signal transduction, causes nevoid BCC syndrome, characterized by susceptibility to BCC (5). Ptch1, a 12-pass transmembrane protein, represses the downstream pathway by inhibiting the seven-transmembrane protein Smo (6–8). This inhibition of Smo is alleviated by the binding of Ptch1 to Hh. Downstream of Smo, Suppressor of fused (Sufu) restrains the activity of the transcriptional effectors of the Hh pathway, the Gli factors (9). In the presence of Hh, Smo activates Gli transcription factors through a process involving modulation of Sufu binding (10).
In vertebrates, Hh signaling is coordinated by the primary cilium, a nonmotile, microtubule-based organelle that projects from the surface of most types of mammalian cells (11, 12). In the absence of Hh pathway activation, Ptch1 localizes to the primary cilium and inhibits Smo ciliary localization (13). In the presence of Hh ligands, Smo accumulates in the cilium and activates the downstream Hh pathway (14). Other Hh pathway components, including Sufu and the Gli transcription factors, also localize to the cilium suggesting that the cilium is the subcellular site at which Smo productively interacts with its downstream targets (9, 15).
Some cancer cells are ciliated, and cilia can block or promote tumorigenesis in the skin and brain, depending on the oncogenes involved (16, 17). BCC caused by misactivation of Smo depends on cilia (17). Given that genetic modulation of ciliogenesis can affect oncogenic Hh signaling, we hypothesized that small molecules that affect ciliogenesis or the movement of Smo to cilia would be valuable tools for dissecting signal transduction and could be starting points for alternative means of targeting Hh pathway-associated cancers.
Inhibitors of several components of the Hh signal transduction pathway can inhibit the growth of some cancers. Robotnikinin is an inhibitor of Sonic hedgehog (Shh) activity, and GANT61, JK184 and HPI4 inhibit Gli transcription factors (18–21), but the majority of identified Hh pathway antagonists inhibit Smo. These include cyclopamine (a natural steroidal alkaloid), Cur61414, Vismodegib/GDC-0449, and LDE225 (22-25). Vismodegib is approved for the treatment of BCC (24). Smo agonists, including the small molecule Smoothened agonist (SAG), have been identified as well (18).
To identify regulators of ciliogenesis and Smo activity, we conducted a high-content screen for compounds that prevent the localization of YFP-tagged Smo to the primary cilium. We sought to identify two classes of small molecules, those that block Smo localization to the primary cilium and those that inhibit ciliogenesis. This screen identified 10 unique SAs, 9 of which inhibit Hh signaling through direct interaction with Smo and 1 of which inhibits Hh signaling but does not interact directly with Smo in a way that interferes with cyclopamine binding. We also identified two ciliogenesis antagonists (CAs) that disrupt ciliogenesis in some cell types. Given that vertebrate Hh signaling and ciliogenesis are intimately linked, both classes allow dissection of the molecular events of Hh signaling.
Results
Development of a High-Throughput Screen for Inhibitors of Smo Ciliary Localization and Ciliogenesis.
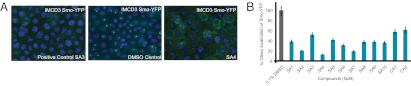
We developed a microscopy-based high throughput screen for small molecules that inhibit ciliogenesis or Smo localization to cilia using IMCD3 cells stably expressing Smo-YFP, selected for their ability to produce long cilia. Because overexpression of Smo causes constitutive ciliary localization, the IMCD3 cell line exhibited constitutive localization of Smo-YFP to the primary cilium (Fig. 1A) (14).
Fig. 1.
Identification of 10 SAs and 2 CAs. (A) Representative high content photomicrographs of the localization of Smo-YFP in IMCD3 cells treated with a positive control compound (SA3), DMSO control, or an experimental compound (SA4). (Smo-YFP, green, Hoechst, blue.) (B) Quantitation of IMCD3 ciliary Smo-YFP from the secondary validation of hits.
Multifactorial experimental design revealed that ciliogenesis was sensitive to cell culture conditions, cell density, and DMSO concentration. IMCD3 ciliation was optimal at a seeding density of 4 × 104 cells/well and a compound incubation time of 16–20 h. Although incubation with 1% DMSO did not cause overt signs of toxicity within 24 h, it unexpectedly increased Smo-YFP localization to cilia; thus, we used 0.1% DMSO.
We imaged cells and analyzed the proportion of cells with cilia marked with Smo-YFP, as described in SI Materials and Methods. In a preliminary trial, we identified a compound, designated SA3, which we used as a positive control during high-throughput screening (Fig. 1A). The optimized assay and analysis conditions had a Z-factor of 0.6, allowing for hit identification with screening in singlicate using 10 μM compound.
Identification of Smo and Ciliogenesis Antagonists.
In an initial trial, we screened 12,250 compounds for their ability to inhibit the localization of Smo-YFP to the primary cilium. We identified 348 compounds that altered ciliary Smo-YFP fluorescence intensity by >2 SDs from the mean. We rescreened these 348 compounds and confirmed that 12 of the compounds abrogated ciliary localization of Smo-YFP in at least 60% of the cells, or 3 SD from the mean (Fig. 1B). By chromatography and mass spectrometry analysis, we confirmed the structures of these 12 compounds, 8 of which are chemically stable and do not contain obvious reactive functionalities. The other four contain potentially reactive functional groups (the α-β unsaturated ketone in SA3, sulfonamide in SA5, thiadiazole in SA10, and pentafluorobenzene sulfonamide in CA2), raising the possibility that these compounds covalently modify their targets (27). However, these compounds were stable in solution and did not induce cell death.
Loss of the ciliary Smo-YFP signal can result from inhibition of Smo-YFP movement to the cilium or loss of the cilium itself. To discriminate between these two mechanisms, we treated IMCD3 Smo-YFP cells with the 12 compounds and assessed the colocalization of Smo-YFP and acetylated tubulin, a ciliary component. Ten of the 12 compounds inhibited the localization of Smo-YFP to the primary cilium, but did not affect acetylated tubulin (Fig. S1A). We named these 10 compounds SA1–10, and the two compounds that disrupted ciliary structure CA1 and CA2 (Fig. S1B). The SA compounds fall into distinct structural clusters (i.e., SA1/2, SA3/4, and SA8/9), suggesting structure–activity relationships. To confirm the specificity of the screen, we identified five control compounds with structural similarity to the SA hits, designated NC1–5, for negative controls (Fig. S1B).
The SA compounds specifically inhibited Smo protein activity but did not block Smo transcription, translation, or the translocation of other proteins to the cilium. None of the SA compounds inhibited the ciliary localization of a GFP-tagged version of somatostatin receptor 3 (SSTR3), indicating that the compounds do not affect constitutive ciliary localization (Fig. S1C). SA treatment did not decrease Smo transcript levels or Smo protein levels (Fig. S1 E and F). Moreover, SA treatment of IMCD3 Smo-YFP cells, ASZ1 cells, and NIH 3T3 cells for 5 d did not induce caspase-3 activation, indicating that the SAs do not induce significant apoptosis (Fig. S1G).
SA1–6 Abrogate Localization of Smo to Primary Cilia of BCC-Like Cells.
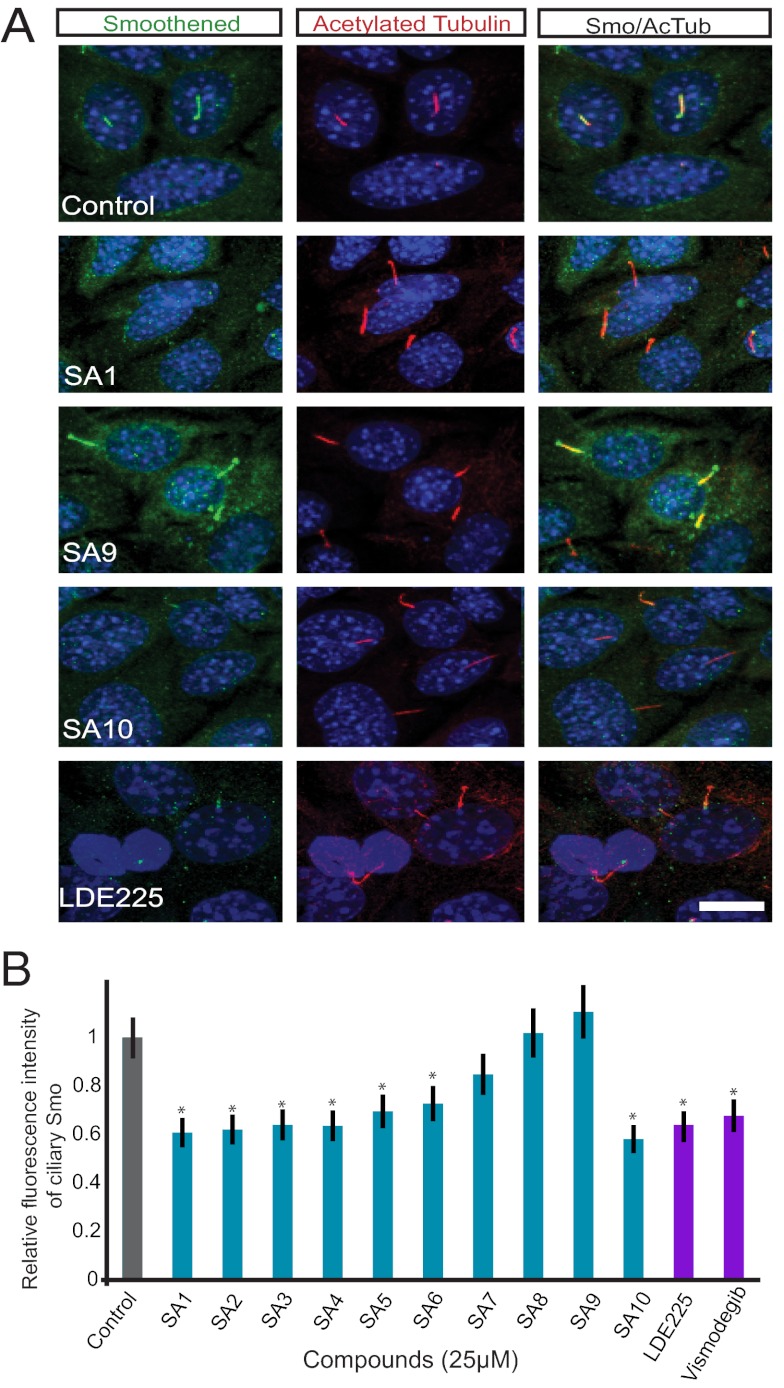
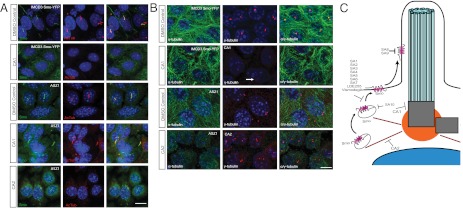
The ability of SA1–10 to inhibit ciliary Smo-YFP localization in IMCD3 cells raised the possibility that these SAs may interfere with the trafficking of endogenous Smo in cancer cells. ASZ1 cells are BCC-like cells derived from tumors that lost Ptch1, display constitutive localization of Smo to the cilium, and constitutively activate the Hh pathway (28, 29). Treatment of ASZ1 cells with 7 of the 10 SA compounds reduced the ciliary localization of endogenous Smo, similar to the clinical Smo antagonist LDE225 (Fig. 2A and Fig. S2); however, SA7–9 did not reduce ciliary Smo in this cell type (Fig. 2B). Given the structural similarity of SA8 and SA9, it is not surprising that these compounds exhibited similar activity.
Fig. 2.
SA1–6 and 10 inhibit the ciliary localization of endogenous Smo in BCC-like cells. (A) Representative immunofluorescent images of decreased Smo localization to cilia in ASZ1 BCC-like cells stained for nuclei (DAPI; blue), Smo (green), and cilia (acetylated tubulin; red). SA1, SA10, and LDE225 but not SA9 decreased localization of ciliary Smo. (Scale bar: 20 μm.) (B) Quantitation of fluorescence intensity of ciliary Smo in ASZ1 cells treated with SAs. Error bars indicate SD throughout figures. Asterisks indicate significance according to the Student t test (P < 0.05) compared with DMSO control.
The inability of SA8 and SA9 to inhibit the ciliary localization of Smo in ASZ1 cells could be related to differences in the levels or trafficking mechanisms of Smo in cancer cells. The Smo antagonist cyclopamine promotes ciliary localization of Smo in many cell types, but inhibits its activity (30). To determine whether SA8 and SA9 act in a cell type-specific fashion similar to cyclopamine, we treated NIH 3T3 cells with SA8 and SA9 in the absence of Hh pathway activators, and found that SA8 and SA9 promoted the localization of endogenous Smo to the primary cilium (Fig. 3A). Thus, SA8 and SA9, like cyclopamine, can promote the ciliary localization of Smo in some cell types.
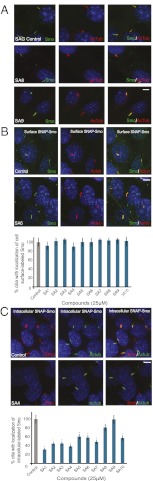
Fig. 3.
Effect of SA compounds on Smo trafficking. SA1–9 compete with BODIPY-cyclopamine for Smo binding. (A) NIH 3T3 cells treated with SAG, SA8, or SA9 and stained for nuclei (DAPI; blue), Smo (green), and cilia (acetylated tubulin; red). 25 μM SA8 or SA9 relocalized Smo to the cilium similar to SAG. (B) SNAP-Smo–expressing MEFs stained for nuclei (DAPI; blue), cell surface SNAP-Smo labeled with SNAP-Surface 488 (green), and cilia (acetylated tubulin; red). 25 μM SA1-10 did not reduce the ciliary localization of cell surface-labeled SNAP-Smo. (C) SNAP-Smo–expressing MEFs stained for nuclei (DAPI; blue), noncell surface SNAP-Smo (labeled by blocking cell surface SNAP-Smo and then labeling with SNAP-Cell TMR Star; red), and cilia (acetylated tubulin; green). Treatment with SA1–7 and 10 reduced the ciliary localization of an intracellular pool of SNAP-Smo. (Scale bar: 20 μm.) Asterisks indicate significance according to the Student t test (P < 0.05) compared with DMSO control.
SA1–7 and SA10 Inhibit Movement of Intracellular, but Not Cell Surface, Smo to Primary Cilia.
Since SA compounds do not reduce Smo levels, we hypothesized that these compounds disrupt Smo trafficking. Smo can be trafficked to the cilium either by lateral transport from the plasma membrane or through an intracellular pathway (22, 31). The kinetics of the two transport events are different, with lateral entry of Smo into the cilium occurring more quickly than entry of the intracellular population of Smo (31).
To determine whether the SAs disrupt trafficking of ciliary Smo at the plasma membrane or intracellular populations, we examined the effects of SA1–10 on cells constitutively expressing a SNAP-tagged version of Smo (SNAP-Smo), Smo bearing an amino-terminal SNAP tag that can be rapidly and covalently labeled with fluorescent small molecules (31). To track the lateral movement of Smo from the plasma membrane to the cilia, we labeled the cell surface population of SNAP-Smo with SNAP-Surface-488, treated with SAG and SA compounds, and assessed whether SNAP-Smo was at the cilia. To track the slower movement of intracellular Smo to the cilia, we blocked the cell surface SNAP-Smo, treated with SAG and SA compounds overnight, and assessed whether labeled intracellular SNAP-Smo was at the cilia immediately after the overnight treatment with SAG and SAs.
SA treatment did not reduce the amount of cell surface SNAP-Smo that moved to the ciliary membrane; however, treatment with SA1–7 and SA10 did reduce the amount of intracellular SNAP-Smo that moved to cilia (Fig. 3 B and C). Consistent with earlier findings, SA8 and SA9 did not inhibit the movement of intracellular Smo to cilia.
SA1–9 Interact Directly with Smo.
To test whether SA compounds antagonize the Hh pathway by interacting directly with Smo, we examined these compounds’ ability to inhibit the binding of BODIPY-cyclopamine, a fluorescent cyclopamine derivative (22). Treatment of Smo-expressing HEK293T cells with 5 μM SA1, SA3, SA4, SA8, and SA9 attenuated BODIPY-cyclopamine binding (Fig. S3A). Higher concentrations (e.g., 10 μM) of SA2, SA5, SA6, and SA7 similarly attenuated BODIPY-cyclopamine binding to Smo (Fig. S3B). SA10, despite its ability to inhibit endogenous ciliary Smo localization, did not displace BODIPY-cyclopamine binding to Smo, indicating that it functions differently than the other SA compounds (Fig. S3C). One possibility is that SA10 may interact with a region of Smo distinct from the cyclopamine site. Alternatively, SA10 may inhibit the machinery that transports Smo to cilia.
SA1–10 Inhibit Smo-Dependent Signal Transduction.
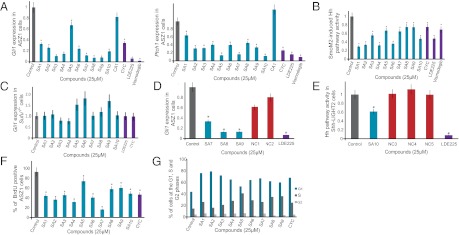
Because Smo localization to cilia is important for vertebrate Hh signaling, we examined whether SA compounds inhibit signal transduction in Hh-responsive Shh-LIGHT2 cells and in two cell lines that display constitutive Hh pathway activation, Ptch1−/− mouse embryonic fibroblasts (MEFs) and ASZ1 cells (19). All the SA compounds inhibited SAG activation of Hh signaling in Shh-LIGHT2 cells at an IC50 of 1–25 μM (22, 32) (Table 1 and Fig. S4). The SA compounds similarly inhibited the constitutive misactivation of the Hh pathway in Ptch1−/− MEFs and ASZ1 cells. Cells lacking Ptch1 activity constitutively express Hh transcriptional targets, including Ptch1 (6). MEFs derived from Ptch1−/− mice also display constitutive localization of ciliary Smo and express β-galactosidase under the control of the Ptch1 promoter (32). SA treatment of Ptch1−/− MEFs suppressed β-galactosidase activity (Fig. S5). As with Shh-LIGHT2 cells, the IC50 of SA compounds in this assay were 1–25 μM (Table 1). We also evaluated the ability of the SA compounds to inhibit expression of Hh pathway target genes, Gli1 and Ptch1, in ASZ1 cells. All 10 SA compounds inhibited expression of Gli1 and Ptch1 in ASZ1 cells (Fig. 4A).
Table 1.
Half maximal inhibitory concentrations (IC50) for Hh pathway (in μM)
| SA1 | SA2 | SA3 | SA4 | SA5 | SA6 | SA7 | SA8 | SA9 | SA10 | CA1 | Cyclopamine | |
| Shh-LIGHT2 cells (Gli-luciferase) | 3.1 | 12 | 1.2 | 0.92 | 5 | 9.2 | 5.8 | 18 | 19 | 5 | >25 | 0.9 |
| Ptch1−/− fibroblast (β-gal) | 3.8 | 9.9 | 20 | 1.3 | 25 | 7.3 | 3.2 | 19 | 1 | 11 | >25 | 0.15 |
Fig. 4.
SA compounds inhibit Hh target gene activation, and SA1–10 reduce proliferation and delay progression through G1 of BCC-like cells. (A) Relative Gli1 and Ptch1 levels in ASZ1 cells treated with 25 μM compound, as measured by qRT-PCR. SA1–10 inhibit the expression of Gli1 and Ptch1 in ASZ1 cells. (B) Relative Gli-luciferase activity of Smo−/− MEFs expressing SmoM2 and treated with 25 μM compound, showing that SA compounds inhibit SmoM2 activity. (C) Quantitation of Gli1 levels in Sufu−/− MEFs treated with 25 μM compound, as measured by qRT-PCR. Sufu is epistatic to SA compound activity. (D) qRT-PCR measurement of relative Gli1 levels in ASZ1 cells treated with 25 μM compound. Compared with their structural analogues SA7–9, NC1 and NC2 are ineffective inhibitors of Gli1 expression. (E) Relative firefly luciferase levels of Shh-LIGHT2 cells treated with SA10 and its structural analogues NC3–5. NC3–5 did not inhibit Hh pathway activity. (F) BrdU incorporation in ASZ1 cells treated with 25 μM compound. SA1–10 reduced cell proliferation in ASZ1 cells. (G) Percentage of cells in each phase of the cell cycle as determined by flow cytometric quantitation of DNA content. ASZ1 cells treated with 25 μM SA1–10 show an increased proportion in G1 phase. Cells treated with SA5 also have an increased proportion in S phase. Data in A–E are averages of quadruplicate measurements; data in F are the average of two independent experiments in duplicate. Asterisks indicate significance according to the Student t test (P < 0.05).
We further assessed whether SA1–10 could inhibit the activity of an oncogenic form of Smo, SmoM2. SmoM2 contains a W535L substitution identified in BCC that causes constitutive ciliary localization and activity (32, 33). We expressed SmoM2-YFP and the Gli-dependent luciferase reporter in Smo−/− MEFs. Treatment with 25 μM SA1–10 inhibited SmoM2-induced Hh pathway activity (Fig. 4B).
Sufu acts downstream of Smo by inhibiting Gli transcription factors and negatively affecting their ability to activate the Hh transcriptional program (34). Consequently, Sufu−/− MEFs exhibit constitutive Hh pathway activity independent of Smo activity (35). To test whether the SA compounds act at the level of Smo, we treated Sufu−/− MEFs with the SA compounds. None of the SA compounds repressed Gli1 expression in these MEFs (Fig. 4C), indicating that Sufu is epistatic to SA function. Thus, it is likely that all 10 SA compounds act at or below Smo, but at or above Sufu.
To provide insight into structure–activity relationships, we investigated the activity of SA7–10 analogues. NC1 and NC2, analogues of SA7–9, were less effective in inhibiting Gli1 expression in ASZ1 cells (Fig. 4D); similarly, NC3–5, analogues of SA10, did not inhibit Hh pathway activity in SAG-treated Shh-LIGHT2 cells (Fig. 4E). The inability of these analogues to inhibit Hh pathway activity demonstrates that small changes in chemotype can strongly reduce activity. Hh signaling shares many features with Wnt signaling (36, 37). To assess whether the SAs also affect Wnt signaling, we tested their ability to inhibit a β-galactosidase reporter under the control of canonical Wnt signals (BATgal). None of the SA compounds inhibited Wnt signaling (Fig. S1H).
SA1–10 Inhibit Proliferation of ASZ1 BCC-Like Cells.
The Hh pathway participates in the regulation of the cell cycle. Hh stimulation of Ptch1 releases cyclin B1, a component of M-phase promoting factor (38). Hh also promotes expression of N-myc and C-myc, which induce cell cycle progression through the induction of CyclinD (38). To determine whether SA1–10 inhibit cell proliferation of Hh pathway-associated cancer cells, we arrested ASZ1 cells in low-serum medium in the presence of 25 μM SA1–10 and 10 μM bromodeoxyuridine (BrdU). After 24 h, we added 10% FBS to promote cell proliferation. SA1–10 reduced BrdU incorporation in ASZ1 cells (Fig. 4F). We further assessed whether SA1–10 inhibit ASZ1 cell proliferation at a particular cell cycle stage by determining the DNA profile of SA1–10-treated ASZ1 cells using flow cytometry. In a nonsynchronous population, SA1–10 delayed the G1-to-S transition, and SA5 also delayed the S-to-G2 transition (Fig. 4G).
CA1 and CA2 Inhibit Ciliogenesis Through Distinct Effects on the Microtubule Cytoskeleton.
In contrast to the SA compounds, CA1 and CA2 blocked cilia formation (Fig. S1D). CA1 disrupted ciliogenesis in IMCD3 cells, but not in ASZ1 and NIH 3T3 cells (Fig. 5A). We recently showed that different cell types have distinct genetic requirements for ciliogenesis, and it is possible that CA1 affects a cell type-specific aspect of ciliogenesis (39, 40). Consistent with the inability of CA1 to inhibit ciliogenesis in ASZ1 and NIH 3T3 cells, CA1 also did not inhibit Hh pathway activity in these cells (Figs. S4 and S5, and Table 1).
Fig. 5.
Mechanisms of CA function. (A) IMCD3 Smo-YFP or ASZ1 cells stained for Smo (green), cilia (acetylated tubulin; red), and nuclei (DAPI; blue), treated with CA1, CA2, or DMSO control. (Scale bar: 20μm.) (B) IMCD3 Smo-YFP or ASZ1 cells treated with 10 μM CA1, CA2, or DMSO control and stained for microtubules (α-tubulin; green), centrosomes (γ-tubulin; red), and nuclei (DAPI; blue). CA1 diminishes γ-tubulin localization to centrosomes, disorganizes microtubules, and leads to the formation of multiple γ-tubulin–positive puncta in some cells (arrow). CA2 disrupts microtubules in ASZ1 cells. (Scale bar: 20 μm.) (C) Model of SA and CA action through five distinct mechanisms. SA1–7 inhibit Hh pathway activity by directly interacting with Smo and inhibiting its ciliary localization. SA8 and SA9 also directly inhibit Smo, but either prevent or stimulate its ciliary localization depending on cell type. SA10 inhibits the movement of Smo to cilia and blocks Hh pathway activation, but does not interact with the cyclopamine site of Smo. CA1 and CA2 inhibit ciliogenesis, with CA1 disrupting centrosome architecture and CA2 disrupting the microtubule cytoskeleton.
CA2 displayed toxicity in IMCD3 and ASZ1 cells at concentrations exceeding 10 μM and in Shh-LIGHT2 and Ptch1−/− MEFs at lower concentrations. As noted above, CA2 contains a potentially reactive pentafluorobenzene sulfonamide group, which may account for its toxicity. At subtoxic concentrations, CA2 disrupted ciliogenesis in both IMCD3 and ASZ1 cells (Fig. 5A). Neither CA1 nor CA2 was able to prevent the binding of BODIPY-cyclopamine to Smo (Fig. S3D). Thus, consistent with the fact that ciliogenesis does not depend on Hh signaling, CA1 and CA2 do not target Smo.
Primary cilia comprise nine doublet microtubules that emanate from basal bodies. To investigate whether CA1 and CA2 disrupts cilia formation by affecting microtubules or basal body organization, we treated IMCD3 Smo-YFP cells with 10 μM CA1 and ASZ1 cells with 10 μM CA2 for 24 h and observed the microtubule cytoskeleton and basal body/centrosome structure. The CA1-treated cells exhibited reduced basal body γ-tubulin, dispersal of γ-tubulin into multiple foci, and a disorganized microtubule cytoskeleton (Fig. 5B). In contrast, CA2-treated cells displayed a loss of microtubules, with no effect on γ-tubulin. Given that both basal bodies and microtubules are essential for ciliogenesis, it is likely that CA1 abrogates ciliogenesis through its effect on basal body structure, whereas CA2 abrogates ciliogenesis by disrupting microtubules.
Discussion
We have developed a high-content, high-throughput screen to identify small molecules that abrogate the ciliary localization of Smo (SA1–10) or disrupt the cilium itself (CA1 and CA2) (Fig. 5C). All of the SA compounds suppress pathological Hh pathway misactivation caused by either a loss of Ptch1 or expression of an oncogenic form of Smo. None of the SAs act epistatic to Sufu, indicating that the SA compounds interfere with Hh signal transduction downstream of Ptch1 and upstream of Sufu.
SA1–6 and SA10 inhibit localization of Smo to cilia, whereas SA8 and SA9 induce the localization of Smo to cilia in ASZ1 cells, but not in IMCD3 cells. SA1–9 inhibit Hh pathway activity through direct interactions with Smo. Thus, SA1–6 share functional characteristics with LDE225 or Vismodegib and SA8 and SA9 share functional characteristics with cyclopamine.
Differential labeling of cell surface and intracellular populations of Smo revealed that SA1–7 and SA10 specifically inhibit the translocation of intracellular Smo to the cilium. The identification of small molecules that discriminate between the movement of cell surface and intracellular Smo to the cilium indicates that the mechanisms underlying these two translocations are distinct (31, 41). Given that SA1–7 and SA10 do not inhibit the movement of cell surface Smo to the cilium but do inhibit Hh signaling in various cell types, full pathway activity may require mobilization of both Smo populations in a nonredundant manner. Alternatively, these results may indicate that the two pools of Smo exert different activities, with the intracellular population having a distinct capacity to activate downstream signaling.
To investigate whether the SA compounds resemble compounds with described biological activities, we used the similarity ensemble approach to screen the SA compounds against the ∼3,000 targets and 500,000 ligands of the ChEMBL database (42–44). This approach predicted that SA6 would be an Smo inhibitor, reflecting its similarity to the 1-amino-4-benzylphthalazine family of Smo inhibitors (45). The dissimilarity of the other SA compounds to known Hh pathway regulators suggests that they affect Smo in distinct ways.
Among the other compounds that bind Smo, Vismodegib, and LDE225 are similar bisaryl amides, whereas cyclopamine is a steroidal alkaloid. Despite their diverse structures, SA1–9, Vismodegib, LDE225, and cyclopamine all compete with BODIPY-cyclopamine for binding to Smo. Thus, SA1–9 expand the broad chemical palette of compounds that can bind Smo and inhibit Hh pathway activity, which may be therapeutically useful given the emergence of resistance to a Smo antagonist (46, 47). Based on the diverse structures of Smo antagonists, one might hypothesize that antagonist activity would not be tightly dependent on structure. However, analogues of SA7–10 exhibit little or no ability to inhibit Hh pathway activity. Thus, the ability of minor changes in side groups to dramatically lower activity indicates that Smo inhibitors are structurally constrained.
SA10, unlike SA1–9, Vismodegib and LDE225, does not compete with cyclopamine for Smo binding; rather, it acts upstream of or at the level of Smo through a distinct mechanism. It is possible that SA10 binds Smo at a site distinct from that of cyclopamine. Alternatively, SA10 may block Hh signaling by inhibiting the transport machinery that moves Smo to cilia.
CAs, CA1 and CA2 inhibit ciliogenesis through distinct mechanisms. CA1 reduces the localization of γ-tubulin to basal bodies, induces the formation of multiple γ-tubulin foci, and disrupts the microtubule cytoskeleton, suggesting that CA1 distorts centrosome composition (48). In contrast, CA2 disrupts cytoplasmic microtubules without affecting basal bodies. Ciliobrevin, a small molecule that inhibits ciliogenesis through the inhibition of cytoplasmic dynein, also disrupts microtubule organization and γ-tubulin localization (49).
In vertebrates, the primary cilium is required for Smo to activate the downstream Hh signaling pathway. Our findings suggest that Smo can be inhibited in three distinct ways: (i) by binding to and preventing the ciliary localization of Smo, (ii) by inactivating ciliary Smo, and (iii) by indirectly blocking Smo movement to cilia.
Because the Hh pathway promotes cancer growth, novel drugs that antagonize Hh signaling components, such as Smo, or disrupt the cilium could prove of therapeutic value. Loss of Ptch1 and constitutive activation of Smo cause BCC and medulloblastoma, but treatment with a single Smo antagonist can fail (50). Therefore, complementary inhibitors, especially those that act through independent mechanisms, may provide synergistic clinical benefit in the treatment of Hh pathway-related cancers.
Materials and Methods
Detailed descriptions of the reagents and protocols used in this study are provided in SI Materials and Methods.
Small Molecules.
SA1-10, CA1-2, and NC3-5 were purchased from ChemDiv. NC1 and NC2 were obtained from the University of California at San Francisco Small Molecules Discovery Center. LDE225 was obtained from Novartis Pharma, and Vismodegib was obtained from Genentech. Cyclopamine (CalBioChem) was used as an additional inhibitor of the Hh pathway. BODIPY-cyclopamine (Medical Isotopes) was used in Smo-binding assays. Recombinant mouse Wnt3a and Dkk-1 (R&D Systems) were used to assess Wnt pathway activity.
Supplementary Material
Acknowledgments
We thank Kurt Thorn and Alice Thwin of the University of California at San Francisco Nikon Imaging Center for microscopy assistance, Helene Faure for the anti-Smo antibody, Jussi Taipale for the Smo−/− MEFs, Brian Hearn for mass spectroscopy assistance, Brian Shoichet for structural insights into the compounds, Raj Rohatgi for SNAP-Smo–expressing fibroblasts, Andrew Kodani for assistance with the immunoblot analysis, and members of the J.F.R. laboratory for critical discussions and reading of the manuscript. This work was funded by National Institutes of Health Grants R01 AR05439 and R21 NS066448, the Burroughs Wellcome Fund, the David and Lucile F. Packard Foundation, and the Sandler Family Supporting Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207170109/-/DCSupplemental.
References
- 1.Keiser MJ, et al. Predicting new molecular targets for known drugs. Nature. 2009;462:175–181. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berman DM, et al. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425:846–851. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- 3.Watkins DN, et al. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature. 2003;422:313–317. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
- 4.Watkins DN, Berman DM, Baylin SB. Hedgehog signaling: Progenitor phenotype in small-cell lung cancer. Cell Cycle. 2003;2:196–198. [PubMed] [Google Scholar]
- 5.Hahn H, et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85:841–851. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 6.Goodrich LV, Johnson RL, Milenkovic L, McMahon JA, Scott MP. Conservation of the hedgehog/patched signaling pathway from flies to mice: Induction of a mouse patched gene by Hedgehog. Genes Dev. 1996;10:301–312. doi: 10.1101/gad.10.3.301. [DOI] [PubMed] [Google Scholar]
- 7.Johnson RL, et al. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science. 1996;272:1668–1671. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- 8.May SR, et al. Loss of the retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli. Dev Biol. 2005;287:378–389. doi: 10.1016/j.ydbio.2005.08.050. [DOI] [PubMed] [Google Scholar]
- 9.Haycraft CJ, et al. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1:e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tukachinsky H, Lopez LV, Salic A. A mechanism for vertebrate Hedgehog signaling: Recruitment to cilia and dissociation of SuFu-Gli protein complexes. J Cell Biol. 2010;191:415–428. doi: 10.1083/jcb.201004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huangfu D, et al. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- 12.Goetz SC, Anderson KV. The primary cilium: A signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 14.Corbit KC, et al. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 15.Liu A, Wang B, Niswander LA. Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development. 2005;132:3103–3111. doi: 10.1242/dev.01894. [DOI] [PubMed] [Google Scholar]
- 16.Han YG, et al. Dual and opposing roles of primary cilia in medulloblastoma development. Nat Med. 2009;15:1062–1065. doi: 10.1038/nm.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong SY, et al. Primary cilia can both mediate and suppress Hedgehog pathway-dependent tumorigenesis. Nat Med. 2009;15:1055–1061. doi: 10.1038/nm.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank-Kamenetsky M, et al. Small-molecule modulators of Hedgehog signaling: Identification and characterization of Smoothened agonists and antagonists. J Biol. 2002;1:10. doi: 10.1186/1475-4924-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hyman JM, et al. Small-molecule inhibitors reveal multiple strategies for Hedgehog pathway blockade. Proc Natl Acad Sci USA. 2009;106:14132–14137. doi: 10.1073/pnas.0907134106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiselyov AS. Targeting the hedgehog signaling pathway with small molecules. Anticancer Agents Med Chem. 2006;6:445–449. doi: 10.2174/187152006778226495. [DOI] [PubMed] [Google Scholar]
- 21.Peng LF, Stanton BZ, Maloof N, Wang X, Schreiber SL. Syntheses of aminoalcohol-derived macrocycles leading to a small-molecule binder to and inhibitor of Sonic Hedgehog. Bioorg Med Chem Lett. 2009;19:6319–6325. doi: 10.1016/j.bmcl.2009.09.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 2002;16:2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Büttner A, et al. Synthesis and biological evaluation of SANT-2 and analogues as inhibitors of the hedgehog signaling pathway. Bioorg Med Chem. 2009;17:4943–4954. doi: 10.1016/j.bmc.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 24.Peukert S, Miller-Moslin K. Small-molecule inhibitors of the hedgehog signaling pathway as cancer therapeutics. ChemMedChem. 2010;5:500–512. doi: 10.1002/cmdc.201000011. [DOI] [PubMed] [Google Scholar]
- 25.Yang H, et al. Converse conformational control of smoothened activity by structurally related small molecules. J Biol Chem. 2009;284:20876–20884. doi: 10.1074/jbc.M807648200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J, et al. Functional genomic screen for modulators of ciliogenesis and cilium length. Nature. 2010;464:1048–1051. doi: 10.1038/nature08895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baell JB, Holloway GA. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 28.Yang ZJ, et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell. 2008;14:135–145. doi: 10.1016/j.ccr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.So PL, et al. Long-term establishment, characterization and manipulation of cell lines from mouse basal cell carcinoma tumors. Exp Dermatol. 2006;15:742–750. doi: 10.1111/j.1600-0625.2006.00465.x. [DOI] [PubMed] [Google Scholar]
- 30.Rohatgi R, Milenkovic L, Corcoran RB, Scott MP. Hedgehog signal transduction by Smoothened: Pharmacologic evidence for a 2-step activation process. Proc Natl Acad Sci USA. 2009;106:3196–3201. doi: 10.1073/pnas.0813373106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milenkovic L, Scott MP, Rohatgi R. Lateral transport of Smoothened from the plasma membrane to the membrane of the cilium. J Cell Biol. 2009;187:365–374. doi: 10.1083/jcb.200907126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taipale J, et al. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- 33.Xie J, et al. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature. 1998;391:90–92. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- 34.Pearse RV, 2nd, Collier LS, Scott MP, Tabin CJ. Vertebrate homologs of Drosophila Suppressor of fused interact with the gli family of transcriptional regulators. Dev Biol. 1999;212:323–336. doi: 10.1006/dbio.1999.9335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Svärd J, et al. Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev Cell. 2006;10:187–197. doi: 10.1016/j.devcel.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 36.Barker N. The canonical Wnt/beta-catenin signalling pathway. Methods Mol Biol. 2008;468:5–15. doi: 10.1007/978-1-59745-249-6_1. [DOI] [PubMed] [Google Scholar]
- 37.Wu D, Pan W. GSK3: A multifaceted kinase in Wnt signaling. Trends Biochem Sci. 2010;35:161–168. doi: 10.1016/j.tibs.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kenney AM, Rowitch DH. Sonic hedgehog promotes G(1) cyclin expression and sustained cell cycle progression in mammalian neuronal precursors. Mol Cell Biol. 2000;20:9055–9067. doi: 10.1128/mcb.20.23.9055-9067.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia-Gonzalo FR, et al. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat Genet. 2011;43:776–784. doi: 10.1038/ng.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dowdle WE, et al. Disruption of a ciliary B9 protein complex causes Meckel syndrome. Am J Hum Genet. 2011;89:94–110. doi: 10.1016/j.ajhg.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y, Zhou Z, Walsh CT, McMahon AP. Selective translocation of intracellular Smoothened to the primary cilium in response to Hedgehog pathway modulation. Proc Natl Acad Sci USA. 2009;106:2623–2628. doi: 10.1073/pnas.0812110106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeGraw AJ, Keiser MJ, Ochocki JD, Shoichet BK, Distefano MD. Prediction and evaluation of protein farnesyltransferase inhibition by commercial drugs. J Med Chem. 2010;53:2464–2471. doi: 10.1021/jm901613f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferreira RS, et al. Complementarity between a docking and a high-throughput screen in discovering new cruzain inhibitors. J Med Chem. 2010;53:4891–4905. doi: 10.1021/jm100488w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kolb P, et al. Structure-based discovery of beta2-adrenergic receptor ligands. Proc Natl Acad Sci USA. 2009;106:6843–6848. doi: 10.1073/pnas.0812657106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mott BT, et al. Identification and optimization of inhibitors of Trypanosomal cysteine proteases: cruzain, rhodesain, and TbCatB. J Med Chem. 2010;53:52–60. doi: 10.1021/jm901069a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller-Moslin K, et al. 1-Amino-4-benzylphthalazines as orally bioavailable smoothened antagonists with antitumor activity. J Med Chem. 2009;52:3954–3968. doi: 10.1021/jm900309j. [DOI] [PubMed] [Google Scholar]
- 47.Yauch RL, et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science. 2009;326:572–574. doi: 10.1126/science.1179386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mikule K, et al. Loss of centrosome integrity induces p38-p53-p21-dependent G1-S arrest. Nat Cell Biol. 2007;9:160–170. doi: 10.1038/ncb1529. [DOI] [PubMed] [Google Scholar]
- 49.Firestone AJ, et al. Small-molecule inhibitors of the AAA+ ATPase motor cytoplasmic dynein. Nature. 2012;484:125–129. doi: 10.1038/nature10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rudin CM, et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med. 2009;361:1173–1178. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.