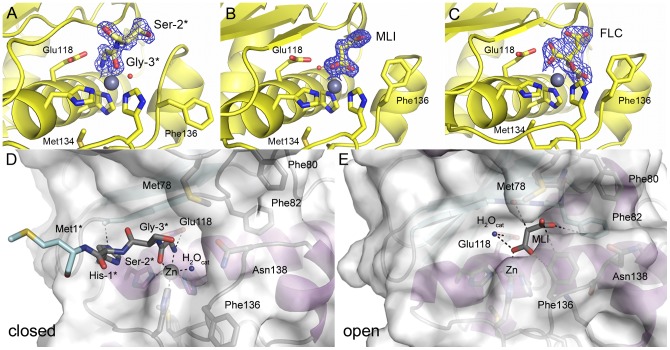
Figure 4. Structural differences between NHis-AfAmzA and nat-AfAmzA.
Simulated annealing Fo–Fc omit map of (A) the N-terminus of a symmetry-related molecule (Gly-3*-Ser-2*) in NHis-AfAmzA, (B) a malonate molecule (MLI) in nat-AfAmzA::malonate and (C) a citrate molecule (FLC) in nat-AfAmzA::citrate, contoured at 2σ level. The maps were generated using phenix.omit_map [37] and converted to the ccp4 format with FFT (V6.1) [32]. Important residues are shown as sticks. The zinc ions and water molecules are shown as grey and red spheres, respectively. (D) Surface representation of NHis-AfAmzA active site cleft with the N-terminus of a symmetry related molecule bound to the catalytic zinc ion (primed site in closed conformation). (E) Surface representation of nat-AfAmzA active site cleft with the zinc-bound malonate molecule (MLI; primed site in open conformation). Important residues and the catalytic water molecule (H2Ocat) are labeled. The surface is transparent to allow a view on the residues involved in zinc ion and ligand binding.