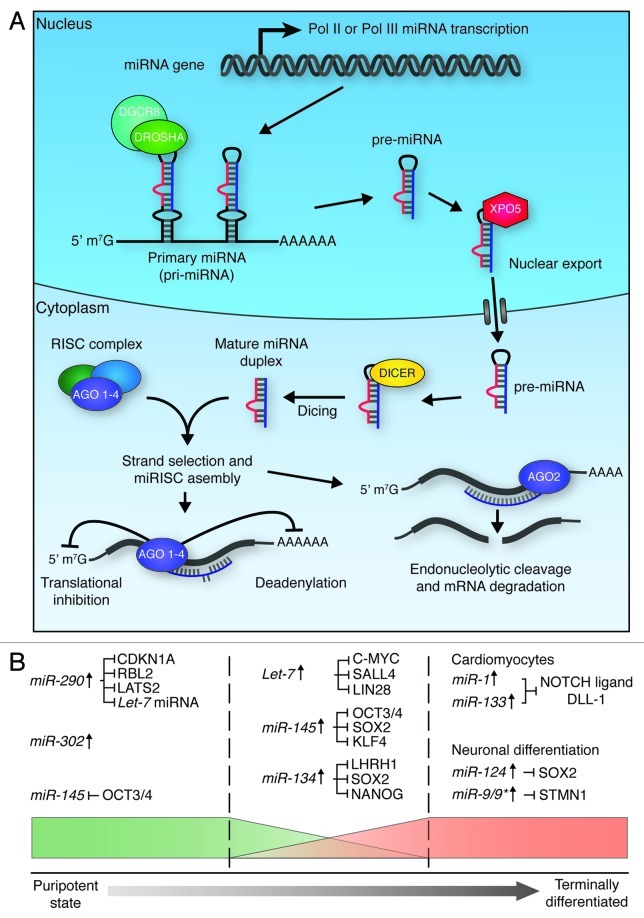
Figure 5. miRNA biogenesis and function in ES cell differentiation. (A) Micro RNA biogenesis. Primary transcripts (pri-miRNAs) are generated through RNA pol II or pol III transcription, which then undergo RNase III cleavage, mediated by the DROSHA/DGCR8 complex, to generate ~70 nt pre-miRNAs. XPO5 exports these pre-miRNAs to the cytoplasm, where they are further cleaved by DICER to generate mature double-stranded RNA duplexes. One strand of these duplexes is then bound by one of four Argonaute proteins (AGO 1–4) to form active RISC complexes, which can modulate gene expression through translational inhibition, or mRNA deadenylation. If the miRNA is perfectly matched to the target sequence, endonucleolytic cleavage of the mRNA transcript can occur through the ‘slicer’ activity of AGO2.139,145 (B) miRNAs play roles in the maintenance of pluripotency (left section), the onset of differentiation (middle section), and the maintenance of terminal differentiation (right section). In order to maintain pluripotency, miRNAs act to promote maintenance of cell cycle progression (CDKN1A, RBL2 and LATS2 inhibition) and de novo DNA methylation (RBL2 inhibition), and suppress factors that promote differentiation (Let-7 inhibition). In order to promote differentiation, miRNAs act to block self-renewal and core pluripotency factor production. miRNAs maintain terminal differentiation by suppressing gene expression associated with other lineages (miR-1 and miR-133), block self-renewal (miR-124) and maintain the state of the specific lineage (miR-9/9*).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.