Abstract
Aims
Nicotine dependence is a highly heritable disorder associated with severe medical morbidity and mortality. Recent meta-analyses have found novel genetic loci associated with cigarettes per day (CPD), a proxy for nicotine dependence. The aim of this paper is to evaluate the importance of phenotype definition (i.e. CPD versus Fagerström Test for Cigarette Dependence (FTCD) score as a measure of nicotine dependence) on genome-wide association studies of nicotine dependence.
Design
Genome-wide association study
Setting
Community sample
Participants
A total of 3,365 subjects who had smoked at least one cigarette were selected from the Study of Addiction: Genetics and Environment (SAGE). Of the participants, 2,267 were European Americans,999 were African Americans.
Measurements
Nicotine dependence defined by FTCD score ≥4, CPD
Findings
The genetic locus most strongly associated with nicotine dependence was rs1451240 on chromosome 8 in the region of CHRNB3 (OR=0.65, p=2.4×10−8). This association was further strengthened in a meta-analysis with a previously published dataset (combined p=6.7 ×10−16, total n=4,200).When CPD was used as an alternate phenotype, the association no longer reached genome-wide significance (β=−0.08, p=0.0007).
Conclusions
Daily cigarette consumption and the Fagerstrom Test for Cigarette Dependence (FTCD) show different associations with polymorphisms in genetic loci.
INTRODUCTION
Tobacco use is one of the leading causes of mortality worldwide. Because most regular smoking occurs in the context of nicotine dependence, nicotine dependence is frequently the focus of studies on tobacco use(1). Among current smokers, approximately 60% are nicotine dependent based on the Fagerström Test for Cigarette Dependence (FTCD), a well-established scale for assessing nicotine dependence(2). Evidence for genetic factors contributing to the risk of smoking behaviors and nicotine dependence is seen in the clustering of heavy smoking and nicotine dependence in families and the similarity of smoking behaviors in genetically identical twins(3–4).
Numerous studies have found an association between nicotine dependence and SNPs in the α5 nicotinic receptor gene, CHRNA5(5–12).To maximize the power to detect genetic variants associated with smoking quantity (as an alternative to nicotine dependence), three large research consortia performed genome-wide association meta-analyses of cigarettes per day (CPD)in a combined sample of over 75,000 subjects (13–15).The strongest association was the variant in CHRNA5, with a combined p-value less than 1 × 10−70. However, several other variants were discovered with genome-wide significance: variants near the nicotinic receptor subunit genes CHRNB3 and CHRNA6 on chromosome 8 (rs6474412, p=1.4 × 10−8,a region previously associated with other nicotine phenotypes(16–20)),variants near the nicotine metabolizing enzyme genes CYP2A6 and CYP2B6 on chromosome 19(rs4105144, p=2.2 × 10−12), and variants in a non-coding region located on chromosome 10q23(rs1329650, p=5.7 × 10−10).
Because genome-wide association studies (GWAS) have stringent p-value requirements, the issue of statistical power is highly relevant. Although most attempts at maximizing power in GWAS focus on increasing sample size as in the above meta-analyses, power can also be increased by reducing phenotypic variance by either increasing precision of measurement or increasing phenotypic homogeneity of the subjects.
Although CPD is the most common phenotypic measurement of smoking behavior, there is strong epidemiological evidence that the number of cigarettes smoked per day varies across cultures and ethnicities. For example, African Americans smoke fewer cigarettes than European Americans(21). However, the FTCD score, ranging from 0 to 10 where CPD can account for a maximum of four levels, defined as 1–10, 11–20, 21–30, or 31 or more, appears to be an invariant measure of nicotine dependence across ethnicities(21). Therefore, we hypothesized that a genome-wide association study with FTCD rather than CPD may have increased power to detect variants associated with nicotine dependence, especially in a multi-ethnic sample.
To clarify the relationship between FTCD-based nicotine dependence and CPD in the context of a genome-wide association study, we defined FTCD-based nicotine dependent cases and non-nicotine dependent controls from the Study of Addiction: Genetics and Environment (SAGE), a multi-ethnic, case-control sample selected for alcohol dependence(22). By including a diverse set of study participants, we have the opportunity to extend our investigation beyond the previous studies in European-Americans, and specifically address the role that phenotype definition plays in genome-wide association studies.
METHODS
Data
This analysis uses a subset of subjects who have ever smoked from the Study of Addiction: Genetics and Environment (SAGE), part of the Gene Environment Association Studies (GENEVA) program of the National Institutes of Health (NIH) Genes, Environment, and Health Initiative(23). For the overall SAGE project, unrelated alcohol dependent cases (N = 1,897) and non-alcohol dependent control subjects (N = 1,937) were selected from three large, complementary datasets: Collaborative Genetic Study of Nicotine Dependence (COGEND), Collaborative Study on the Genetics of Alcoholism (COGA), and Family Study of Cocaine Dependence (FSCD). Characteristics of the individual datasets are given in supplementary Table 3. The Institutional Review Board at each contributing institution reviewed and approved the protocols for genetic studies of substance dependence under which all subjects were recruited. Subjects provided informed consent for genetic studies and agreed to allow their genetic and phenotypic information to be shared with qualified investigators through NIH repositories. For each of the three studies, we describe the sampling schemes used to recruit subjects and select for genotyping.
Collaborative Genetic Study of Nicotine Dependence (COGEND)
COGEND was designed as a community based case–control family study of nicotine dependence. The COGEND ascertainment protocol identified current smokers with nicotine dependence defined by an FTCD score ≥4 (maximum score of 10); non-nicotine dependent subjects who had smoked at least 100 cigarettes and had a lifetime FTCD score of zero were also recruited. All subjects were ascertained from Detroit and St. Louis. Approximately 53,000 subjects were screened by telephone, 2,800 were personally interviewed, and nearly 2,700 donated blood samples for genetic studies. The COGEND study contributed 275 nicotine dependent cases and 1,082 non-nicotine dependent smoking controls to this nicotine dependence genetic analyses. Of these, 125 nicotine-dependent cases and 706 non-nicotine dependent controls overlap with the samples used in Bierut et al.2007, and Saccone et al.2007(5–6).
Collaborative Study on the Genetics of Alcoholism (COGA)
A case control series of unrelated individuals was selected from over 8,000 subjects who participated in the genetic arm of COGA. COGA systematically recruited subjects from participating centers in Hartford, Connecticut; Indianapolis, Indiana; Iowa City, Iowa; New York City, New York; San Diego, California; St. Louis, Missouri, and Washington DC. For SAGE, cases met life-time criteria for DSM-IV alcohol dependence; the majority of cases were recruited from alcoholism treatment centers. Control subjects, biologically unrelated to cases, were individuals who consumed alcohol, but never experienced any significant alcohol or drug related problems, as reported on the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA). The COGA study contributed 649 nicotine dependent cases and 553 non-nicotine dependent smoking control subjects to the subsequent nicotine dependence genetic analyses.
Family Study of Cocaine Dependence (FSCD)
Cocaine dependent cases were systematically recruited from chemical dependency treatment units in the greater St. Louis metropolitan area. Community based control subjects were identified through the Missouri Family Registry and matched by age, race, gender, and residential zip code. Controls were biologically unrelated individuals from the same communities who consumed alcohol, but had no lifetime history of dependence on any substance. FSCD contributed 370 nicotine dependent cases and 436 non-nicotine dependent smoking control subjects.
Nicotine Dependence and Smoking Phenotypes
We used several approaches to define the most appropriate phenotype for the genetic association analysis of smoking. First, to enhance sample homogeneity, we eliminated 143 individuals who had substance dependence diagnoses other than nicotine dependence, alcohol dependence, alcohol and cocaine dependence, or other substance abuse (except nicotine dependence). These 143 subjects are labeled as “Other” in the SAGE files available through the database of Genotypes and Phenotypes (dbGaP; accession number phs000092.v1.p1).
We included all subjects who had ever smoked a cigarette. Cases were defined with a Fagerström Test for Cigarette Dependence (FTCD) score of 4 or more, and controls had an FTCD score ≤ 3, based on the dichotomous nicotine dependence phenotype used for COGEND. For our case definition, we also included 100 individuals who smoked on average more than a pack a day, but had a missing FTCD score. This is consistent with previous research that found that most individuals who smoke more than 20 cigarettes a day have an FTCD score of 4 or greater(9). The final sample for association testing contains 1,294 nicotine dependent cases and 2,071 non-dependent controls who have smoked at least one cigarette (Table 1).
Table 1.
Characteristics of the sample.
| Nicotine Dependent Smokers | Controls | |||||
|---|---|---|---|---|---|---|
| EA | AA | Total | EA | AA | Total | |
| n=828 | n=466 | n=1,294 | n=1,538 | n=533 | n=2,071 | |
| no comorbid diagnosis | 9% | 6% | 8% | 73% | 21% | 71% |
| alcohol dependence | 91% | 94% | 41% | 27% | 35% | 30% |
| cocaine dependence | 43% | 66% | 51% | 8% | 20% | 11% |
| cigarettes per day | ||||||
| 0–10 | 4% | 24% | 11% | 88% | 90% | 89% |
| 11–20 | 39% | 53% | 44% | 11% | 9% | 10% |
| 21–30 | 25% | 9% | 19% | 1% | 0% | 1% |
| ≥31 | 32% | 14% | 25% | 0% | 0% | 0% |
| Age | ||||||
| mean age | 40 | 40 | 40 | 38 | 40 | 39 |
| < 35 | 29% | 18% | 25% | 35% | 20% | 31% |
| 35–39 | 23% | 27% | 24% | 23% | 25% | 24% |
| 40–44 | 23% | 32% | 26% | 25% | 32% | 27% |
| ≥45 | 25% | 23% | 24% | 16% | 23% | 18% |
| male | 60% | 55% | 58% | 36% | 47% | 39% |
| female | 40% | 45% | 42% | 63% | 53% | 61% |
| Income < $20,000 | 15% | 37% | 21% | 3% | 18% | 5% |
| No High School Degree | 14% | 33% | 23% | 3% | 13% | 7% |
A relatively large proportion of individuals in this sample have a diagnosis of alcohol dependence and/or cocaine dependence because the COGA and FSCD studies were designed to examine these disorders. This reflects the elevated rates of nicotine dependence in individuals with comorbid substance dependence conditions.
CPD is an alternative phenotype for smoking behavior that has been studied in previous GWAS. To evaluate CPD, a four point ordinal scale was created: at most 10 cigarettes daily, 11 to 20 cigarettes daily, 21 to 30 cigarettes daily, and more than 30 cigarettes daily. This phenotype has been used in other studies(10). We used this to further examine our top GWAS finding.
Genotyping and Data Cleaning
As part of GENEVA, DNA samples were genotyped on the Illumina Human 1M-Duo beadchip by the Center for Inherited Disease Research (CIDR) at Johns Hopkins University.
The Illumina 1M –Duo array has a total of 1,072,820 probes, of which 23,812 are “intensity-only,” leaving 1,049,008 probes as SNP assays. These SNP assays demonstrate excellent data quality—95% of SNPs have a missing call rate< 1.4% and the median of the missing call rate is 0.05%. A thorough data cleaning procedure was applied to ensure the highest possible data quality, including the use of HapMap controls, detection of gender and chromosomal anomalies, hidden relatedness, population structure, missing call rates, batch effects, Mendelian error detection, duplication error detection, and Hardy-Weinberg equilibrium(23). Of the 1,049,008 SNPs, 948,658 SNPs passed data cleaning procedures. Further details are provided in the comprehensive data cleaning report posted on the GENEVA website http://www.genevastudy.org/docs/GENEVA_Alcohol_QC_report_8Oct2008.pdf).
Population Stratification
The composition of the samples in terms of self-identified ethnicity was 2,267 European Americans (self-reported “white”), 99 Hispanic Americans, and 999 African Americans (self-reported “black”). Subjects identified as both African American and Hispanic were labeled as African Americans.
We used the software package EIGENSTRAT (24)to calculate principal components reflecting continuous variation in allele frequencies, representing ancestral differences in subjects. Two principal components were identified. The first distinguished African American participants from European American participants, and the second distinguished Hispanic from non-Hispanic subjects. These scores were included to control for effects of population stratification. In addition, we used self-reported ethnicity (European American, African American or Hispanic) as a categorical variable and compared results with those using the first two principal components.
Statistical Analyses
Two genome-wide association analyses were conducted in PLINK (25). The first used logistic regression with nicotine dependence as the dependent variable, and the second used linear regression with CPD as the dependent variable. Genotypes were coded log-additively (0, 1, 2 copies of the minor allele). Covariates representing sex, age (defined, using quartiles, as 3 indicator variables representing 34 years and younger (reference), 35–39 years, 40–44 years, and 45 years and older), self-reported ethnicity, and alcohol and cocaine dependence (the diagnoses used to ascertain subjects for the original COGA and FSCD studies) were included.
The QQ-Plot of the association between nicotine dependence (FTCD ≥4) and the 948,658 SNPs may be seen in Figure 1 of the supplementary material. The lambda value is 1.02, reflecting adequate control of population stratification using self-reported ethnicity. The Manhattan plots for the two analyses are shown in Figure 2 of the supplementary material.
To evaluate the robustness of the findings, we analyzed the association between rs1451240 and several smoking phenotypes (cpd and FTCD, coded as a continuous variable in linear regression, dichotomous variable in logistic regression, and ordinal variable in ordered logistic regression). In addition, we modified the inclusion criteria to include only individuals who smoked regularly to determine whether this changed the results.
We performed a meta-analysis of the independent subset of this study using Metal (26–27). The primary sample we used was the 2,590 subjects that were not included in the previous GWAS of nicotine dependence(5). These results were combined with the published odds ratio and corresponding statistics from the association study in 1,610 subjects from Saccone et al.(28).
We graphically evaluated linkage disequilibrium and GWAS p-values using the software WGA viewer (http://people.genome.duke.edu/~dg48/WGAViewer/)(29).
Population-based analysis of smoking phenotypes
To clarify discrepancies between results obtained using FTCD-based nicotine dependence and results obtained with CPD, we compared FTCD to CPD in a population-based sample. As part of recruitment to the COGEND study, subjects were randomly selected from the St. Louis region using the Missouri Family Registry, sent a letter, and called on the phone(3, 6). To evaluate the relationship between the phenotypes of CPD and the FTCD score, of the 28,658 subjects who completed the telephone screening, we selected the 14,343 subjects who had smoked at least 100 cigarettes in their lifetime and calculated the correlation between FTCD and CPD. We also calculated the polychoric correlations between the FTCD score components and CPD.
RESULTS
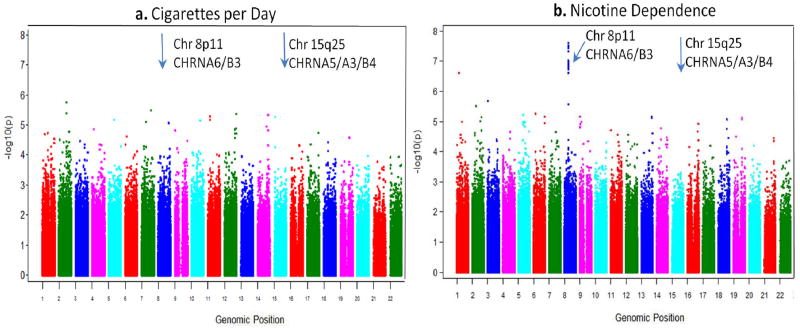
The region most strongly associated with nicotine dependence in this study is represented by 14SNPs in a 40kb region on chromosome 8 (Table 2), with a single bin reaching genome-wide significance (p < 5 × 10−8). The most significant SNP is rs1451240 (OR=0.65,p = 2.4 × 10−8), and this SNP tags a bin on chromosome 8 including part of CHRNB3 in both African Americans and European Americans (supplemental Figures 2 & 3). Of interest, the SNP most strongly associated with nicotine dependence in previous studies, rs16969968in CHRNA5 on chromosome 15, had an odds ratio consistent with published studies (OR=1.31), but the p-value was not genome-wide significant (p=6.2 × 10−4). In contrast with the results from nicotine dependence, the GWAS using CPD as the dependent variable does not find any SNP to be significantly associated at a genome-wide level(Figure 1).
Table 2.
The top 10 associated SNPs from the GWAS analysis of nicotine dependence
| SNP | Chr | Position (Ensembl 56) | Locus and Context | Test Allele | EA Cases | EA Cntrls | EA OR (95% CI) | AA Cases | AA Cntrls | AA OR (95% CI) | Hisp Cases | Hisp Cntrls | Full Dataset Adjusted OR (95% CI) | p |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1451240 | 8 | 42,546,711 | CHRNB3, Intergenic | A | 0.18 | 0.23 | 0.65 (0.53, 0.79) | 0.62 | 0.68 | 0.64 (0.50, 0.81) | 0.25 | 0.31 | 0.65(0.56,0.76) | 2.44E-08 |
| rs4736835 | 8 | 42,547,033 | CHRNB3, Intergenic | T | 0.19 | 0.23 | 0.65 (0.53, 0.80) | 0.62 | 0.68 | 0.64 (0.51, 0.82) | 0.25 | 0.31 | 0.65(0.56,0.76) | 2.99E-08 |
| rs10958725 | 8 | 42,524,584 | CHRNB3, Intergenic | T | 0.18 | 0.23 | 0.65 (0.53, 0.80) | 0.66 | 0.71 | 0.62 (0.48, 0.79) | 0.25 | 0.31 | 0.65(0.56,0.76) | 3.06E-08 |
| rs6474413 | 8 | 42,551,064 | CHRNB3, Upstream | C | 0.19 | 0.23 | 0.65 (0.53, 0.79) | 0.66 | 0.71 | 0.63 (0.50, 0.81) | 0.27 | 0.31 | 0.65(0.56,0.76) | 3.62E-08 |
| rs1955185 | 8 | 42,549,647 | CHRNB3, Upstream | G | 0.19 | 0.23 | 0.65 (0.53, 0.80) | 0.66 | 0.71 | 0.64 (0.50, 0.82) | 0.27 | 0.31 | 0.65(0.56,0.76) | 4.64E-08 |
| rs4950 | 8 | 42,552,633 | CHRNB3,5prime Utr | C | 0.19 | 0.23 | 0.64 (0.52, 0.79) | 0.7 | 0.75 | 0.65 (0.50, 0.85) | 0.28 | 0.33 | 0.65(0.56,0.76) | 9.50E-08 |
| rs7004381 | 8 | 42,551,161 | CHRNB3, Upstream | A | 0.19 | 0.23 | 0.65 (0.53, 0.80) | 0.57 | 0.62 | 0.68 (0.54, 0.86) | 0.25 | 0.31 | 0.67(0.57,0.77) | 9.93E-08 |
| rs13280604 | 8 | 42,559,586 | CHRNB3, Intronic | G | 0.19 | 0.23 | 0.65 (0.53, 0.80) | 0.7 | 0.74 | 0.65 (0.50, 0.84) | 0.28 | 0.33 | 0.66(0.56,0.77) | 1.04E-07 |
| rs10958726 | 8 | 42,535,909 | CHRNB3, Intergenic | G | 0.18 | 0.23 | 0.65 (0.53, 0.80) | 0.57 | 0.63 | 0.68 (0.54, 0.86) | 0.25 | 0.3 | 0.67(0.57,0.77) | 1.15E-07 |
| rs13273442 | 8 | 42,544,017 | CHRNB3, Intergenic | A | 0.19 | 0.23 | 0.65 (0.53, 0.80) | 0.57 | 0.62 | 0.69 (0.55, 0.87) | 0.25 | 0.31 | 0.67(0.58,0.78) | 1.38E-07 |
Figure 1.
Manhattan plot of p-values from multivariate logistic regression models testing for association with cigarettes per day (a) and nicotine dependence (b).
The CHRNB3 region of chromosome 8in general, and this signal in particular, has been previously associated with CPD, but, of the many genome-wide association studies using CPD as the primary phenotype(8, 10, 15, 30), the only previously published genome-wide significant association with this region has been in a large meta-analysis including over 75,000 subjects(15). Specifically, rs1451240 has an r2 of 1.0 (based on 1000 genome pilot 1 data, CEU) with the two chromosome 8 SNPs published in the meta-analysis. To clarify the difference between our study of only 3,365 subjects and the large meta-analysis, we examined the effects of phenotype definition, ethnicity, and comorbid diagnoses on the association with this SNP.
There are two primary differences between the FTCD definition of nicotine dependence and CPD: (1) nicotine dependence is based on a 10 point FTCD scale computed from 6 items including CPD, and (2) nicotine dependence is a dichotomous variable whereas CPD is an ordinal variable. Therefore we created four phenotypes for evaluation: dichotomous nicotine dependence, dichotomous CPD, ordinal FTCD score, ordinal CPD. Using each of these phenotypes, we looked at the association with rs1451240 in multiple stratifications of the data: gender, ethnicity, age, and comorbid substance dependence (Table 3).
Table 3.
Stratified analysis of association between smoking behavior and the CHRNB3 region on chromosome 8 tagged by rs1451240. Interaction p-values correspond to the test that the SNP*strata analysis in the full sample is statistically significant, i.e. the association between SNP and phenotype differs across strata (df = number of strata minus 1).
| association between smoking & rs1451240 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| dichotomous traits | ordinal traits | ||||||||
|
| |||||||||
| FTCD≤3 vs FTCD ≥4 | CPD≤20 vs CPD>20 | FTCD | CPD | ||||||
| n | OR | p | OR | p | β | p | β | p | |
| Full Sample | 3,365 | 0.65 | 2.E-08 | 0.73 | 0.0004 | −0.5 | 0.0004 | −0.08 | 0.0004 |
|
| |||||||||
| stratified analyses | - | ||||||||
|
| |||||||||
| male | 1,555 | 0.69 | 0.0004 | 0.75 | 0.01 | −0.65 | 0.002 | −0.1 | 0.006 |
| female | 1,810 | 0.60 | 0.00002 | 0.72 | 0.02 | −0.35 | 0.04 | −0.06 | 0.04 |
| interaction p-value | 0.17 | 0.09 | 0.32 | 0.0001 | |||||
|
| |||||||||
| European Americans | 2,366 | 0.64 | 1.E-05 | 0.65 | 8.E-05 | −0.54 | 0.001 | −0.1 | 0.0005 |
| African Americans | 999 | 0.64 | 3.E-04 | 0.91 | 6.E-01 | −0.37 | 0.08 | −0.03 | 0.36 |
| interaction p-value | 0.8 | 0.11 | 0.55 | 0.16 | |||||
|
| |||||||||
| age quartile 1 (<34) | 972 | 0.79 | 0.1 | 0.92 | 0.7 | −0.22 | 0.4 | −0.02 | 0.6 |
| age quartile 2 (35–39) | 809 | 0.53 | 6.E-06 | 0.91 | 0.6 | −0.73 | 0.005 | −0.06 | 0.2 |
| age quartile 3 (40–44) | 899 | 0.57 | 0.0005 | 0.72 | 0.06 | −0.43 | 0.06 | −0.1 | 0.04 |
| age quartile 4 (≥45) | 685 | 0.71 | 0.03 | 0.51 | 0.0001 | −0.52 | 0.14 | −0.16 | 0.004 |
| interaction p-value | 0.001 | 0.004 | 0.7 | 0.004 | |||||
|
| |||||||||
| No comorbid substance dependence | 1,564 | 0.68 | 0.03 | 0.59 | 0.09 | −0.1 | 0.6 | −0.02 | 0.25 |
| Alcohol Dependence | 916 | 0.63 | 0.0001 | 0.62 | 0.0005 | −1 | 0.0006 | −0.2 | 0.0004 |
| Alcohol & Cocaine dependence | 885 | 0.67 | 0.0020 | 0.9 | 0.38 | −0.6 | 0.01 | −0.04 | 0.4 |
| interaction p-value | 0.41 | 0.3 | 0.8 | 3.71E-08 | |||||
A substantial decrease in power is noted both with the conversion of nicotine dependence to a dichotomous CPD phenotype, and with the conversion of nicotine dependence to an ordinal FTCD variable. This loss of power is consistent across strata: within nearly every strata of gender, ethnicity, age, and comorbid diagnosis, the strongest association with rs1451240 is seen in the nicotine dependence phenotype. Indeed, tests of proportional odds for both CPD and FTCD scores indicate that there is a threshold effect (p<0.0001 in both cases). Further, varying the definitions of cases and controls does not seem to impact the results (supplemental Tables 1 and 2).
There does not appear to be an effect of gender or comorbid substance dependence on the strength of the association. However, the strength of this difference varies across ethnicity and age. Specifically, despite the fact that the FTCD based definition of nicotine dependence appears to have an equivalent relationship to rs1451240 across ethnicities, the relationship between this SNP and CPD is much weaker in African Americans (β=−0.03, p=0.35), than in European Americans (β=−0.11, p=0.0005). This supports theories that nicotine dependence in African Americans is not fully captured by CPD, likely related to observations that nicotine-dependent African Americans smoke fewer CPD than European Americans(21).The equivalence of these odds ratios across ethnicities is striking especially given that the allele frequencies differ widely in the two groups. For example, the “A” allele of rs1451240 has frequencies of approximately 25% in European American controls and 70% in African American controls. The phenomenon of similar ORs across ethnicities despite different allele frequencies is considered further evidence of a true biological association(31).
To clarify the relationship between CPD and FTCD-based nicotine dependence, we evaluated the correlation between total FTCD score and CPD. In a population sample of subjects from Missouri who had smoked at least 100 cigarettes, the correlation between FTCD and CPD was 0.81 in European Americans (n=11,312) and 0.71 in African Americans (n=3,031). The FTCD items related to early morning smoking had the lowest tetrachoric correlations with CPD both in European Americans (0.35) and African Americans (0.32): item 3: Which cigarette would you hate most to give up (1st AM cigarette), and item 5: Do you smoke more frequently during the first hours after waking than during the rest of the day. In a population sample of Missouri smokers, the correlation between CPD and early morning smoking was 0.69 in EA (n=11,286) and 0.58 in AA (n=3,028). FTCD has been previously described as a two-dimensional phenotype characterized by (1) CPD and (2) early morning smoking(32). The association between early morning smoking and rs1451240 is given in supplemental Table 2. These results suggest that studies using CPD as a phenotype may be missing this important component of nicotine dependence. Furthermore, this discrepancy appears to be of particular relevance in populations of African descent.
Our analysis of the association between nicotine dependence and the SNP rs1451240was combined into a meta-analysis with an independent study of nicotine dependence(6). The previously published study had some subjects that overlapped with the current study. Using the published odds ratio from this study, and eliminating the overlapping subjects from our current study, we computed a meta-analysis p-value of 6.7×10−16 (n=4,200 subjects), further evidence that this association is, indeed, real.
DISCUSSION
We compared two genome-wide association studies of smoking behavior to evaluate the importance of phenotype in genome-wide association studies. We found SNPs on chromosome 8 in the region of CHRNB3 that reached genome-wide significance in their association with nicotine dependence, but did not reach genome-wide significance in the GWAS using CPD as a dependent variable. Interestingly, the association was stronger in our combined sample of 4,200(p=6.7×10−16), than in the meta-analysis of CPD with a combined sample of over 75,000 subjects (p=1.3×10−8)(15).We attribute this discrepancy to the use of an FTCD-based definition of nicotine dependence rather than CPD.
It is important to note that although the correlation between FTCD and CPD is relatively high, the slight change of phenotype from FTCD-based nicotine dependence to CPD changes the results of the study. This has implications in other fields of medicine, implying that a small change in phenotype may expose previously undiscovered variants, and these variants may have specific roles in distinguishing differences between the two phenotypes. Rather than focusing only on increasing the sample size via meta-analyses, this study shows that samples with precise phenotypes may find previously undiscovered variants by conducting association studies using secondary phenotypes.
We specifically examined the relationship between FTCD and CPD to clarify the discrepancy between our FTCD-based results and CPD-based results. Specifically, FTCD includes measures for early morning smoking that are not well-captured by CPD. The difference between these phenotypes may be explained by the contrast in African Americans: although the odds ratios for the SNP using the FTCD-based definition of nicotine dependence are identical in European Americans and African Americans, the effect size for the regression onto CPD is subjectively smaller in African Americans than in European Americans(although not statistically significantly smaller).The inconsistent measurement of CPD as compared to FTCD has been previously described in the literature(21).
It is interesting to note that the FTCD phenotype is strongest as a dichotomous variable, and the highly significant test for proportional odds indicates that the relationship between nicotine dependence and this region is a threshold phenomenon. This suggests that the relationship between CHRNB3 and smoking behavior may be more related to specific component of nicotine dependence rather than smoking quantity.
A second characteristic of our dataset that differs from previously published studies is the enrichment of our sample for substance dependence. Although we did not see a statistical interaction between comorbid diagnosis and the genetic association, our sample was primarily ascertained for substance dependence (alcohol and cocaine). Of interest, the relationship between this region and alcohol dependence has been noted in the literature(22, 33–34). This highlights the complex relationship between comorbid substance use disorders and genetic susceptibilities. Further, although this analysis shows a GWAS-significant association with FTCD-based nicotine dependence that was also seen in a large meta-analysis using CPD as the primary phenotype, it would be interesting to examine the association with this variant in other datasets that have measured FTCD.
Our study highlights a variant associated with nicotine dependence that is more strongly associated with an FTCD-based definition of nicotine dependence than the more common phenotype of CPD. This serves as a striking example of how small changes in phenotype can expose new genetic variants associated with disease.
Supplementary Material
Acknowledgments
The authors thank the following people from Washington University who contributed without compensation: Sherri Fisher for her assistance in data collection and editing of the manuscript, and Hilary Davidson for her assistance with the manuscript.
The Collaborative Genetic Study of Nicotine Dependence (COGEND) project is a collaborative research group and part of the NIDA Genetics Consortium. Subject collection was supported by NIH grant P01 CA089392 (L.J. Bierut) from the National Cancer Institute. Phenotypic and genotypic data are stored in the NIDA Center for Genetic Studies (NCGS) at http://zork.wustl.edu/ under NIDA Contract HHSN271200477451C (J. Tischfield and J. Rice). Lead investigators directing data collection are L.J. Bierut, N. Breslau, D. Hatsukami, and E.O. Johnson. The authors thank Heidi Kromrei and Tracey Richmond for their assistance in data collection.
Additional funding was provided by NIH grants UL1RR024992, K02DA021237 and T32MH014677. John P. Rice, Scott Saccone, Alison Goate and Laura J. Bierut are listed as inventors on Issued U.S. Patent 8,080,371, “Markers for Addiction” covering the use of certain SNPs in determining the diagnosis, prognosis, and treatment of addiction. Laura J. Bierut acted as a consultant for Pfizer in 2008.
Footnotes
Declaration of Interest: Funding support for the Study of Addiction: Genetics and Environment (SAGE) was provided through the NIH Genes, Environment and Health Initiative [GEI] (U01 HG004422). SAGE is one of the genome-wide association studies funded as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for collection of datasets and samples was provided by the Collaborative Study on the Genetics of Alcoholism (COGA; U10 AA008401), the Collaborative Genetic Study of Nicotine Dependence (COGEND; P01 CA089392), and the Family Study of Cocaine Dependence (FSCD; R01 DA013423, R01 DA019963). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438), the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Drug Abuse, and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C).
References
- 1.Fiore MC, Jaen CR, Baker TB, Bailey WC, Benowitz NL, Curry SJ, et al. Treating Tobacco Use and Dependence: 2008 Update. U.S. Department of Health and Human Services; 2008. [Google Scholar]
- 2.Heatherton TF, Kozlowski LT, Frecker RC, Fagerstrom KO. The Fagerstrom Test for Nicotine Dependence: a revision of the Fagerstrom Tolerance Questionnaire. Br J Addict. 1991 Sep;86(9):1119–27. doi: 10.1111/j.1360-0443.1991.tb01879.x. [DOI] [PubMed] [Google Scholar]
- 3.Bierut LJ, Dinwiddie SH, Begleiter H, Crowe RR, Hesselbrock V, Nurnberger JI, Jr, et al. Familial transmission of substance dependence: alcohol, marijuana, cocaine, and habitual smoking: a report from the Collaborative Study on the Genetics of Alcoholism. Arch Gen Psychiatry. 1998 Nov;55(11):982–8. doi: 10.1001/archpsyc.55.11.982. [DOI] [PubMed] [Google Scholar]
- 4.Li MD. The genetics of nicotine dependence. Curr Psychiatry Rep. 2006 Apr;8(2):158–64. doi: 10.1007/s11920-006-0016-0. [DOI] [PubMed] [Google Scholar]
- 5.Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF, et al. Novelgenes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007 Jan 1;16(1):24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007 Jan 1;16(1):36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008 May;40(5):616–22. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berrettini W, Yuan X, Tozzi F, Song K, Francks C, Chilcoat H, et al. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry. 2008 Apr;13(4):368–73. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stevens VL, Bierut LJ, Talbot JT, Wang JC, Sun J, Hinrichs AL, et al. Nicotinic receptor gene variants influence susceptibility to heavy smoking. Cancer Epidemiol Biomarkers Prev. 2008 Dec;17(12):3517–25. doi: 10.1158/1055-9965.EPI-08-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008 Apr 3;452(7187):638–42. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiss RB, Baker TB, Cannon DS, von Niederhausern A, Dunn DM, Matsunami N, et al. A candidate gene approach identifies the CHRNA5-A3-B4 region as a risk factor for age-dependent nicotine addiction. PLoS Genet. 2008 Jul;4(7):e1000125. doi: 10.1371/journal.pgen.1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saccone NL, Culverhouse RC, Schwantes-An TH, Cannon DS, Chen X, Cichon S, et al. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS Genet. 2010;6(8) doi: 10.1371/journal.pgen.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heath AC, Madden PA, Bucholz KK, Dinwiddie SH, Slutske WS, Bierut LJ, et al. Genetic differences in alcohol sensitivity and the inheritance of alcoholism risk. Psychol Med. 1999 Sep;29(5):1069–81. doi: 10.1017/s0033291799008909. [DOI] [PubMed] [Google Scholar]
- 14.Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010 May;42(5):436–40. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010 May;42(5):448–53. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehringer MA, McQueen MB, Hoft NR, Saccone NL, Stitzel JA, Wang JC, et al. Association of CHRN genes with “dizziness” to tobacco. Am J Med Genet B Neuropsychiatr Genet. 2009 Sep 16; doi: 10.1002/ajmg.b.31027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenbaum L, Kanyas K, Karni O, Merbl Y, Olender T, Horowitz A, et al. Why do young women smoke? I. Direct and interactive effects of environment, psychological characteristics and nicotinic cholinergic receptor genes. Mol Psychiatry [Research Support, Non-US Gov’t] 2006 Mar;11(3):312–22. 223. doi: 10.1038/sj.mp.4001774. [DOI] [PubMed] [Google Scholar]
- 18.Hoft NR, Corley RP, McQueen MB, Schlaepfer IR, Huizinga D, Ehringer MA. Genetic association of the CHRNA6 and CHRNB3 genes with tobacco dependence in a nationally representative sample. Neuropsychopharmacology [Research Support, NIH, Extramural] 2009 Feb;34(3):698–706. doi: 10.1038/npp.2008.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet [Research Support, NIH, Extramural] 2007 Jan 1;16(1):36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeiger JS, Haberstick BC, Schlaepfer I, Collins AC, Corley RP, Crowley TJ, et al. The neuronal nicotinic receptor subunit genes (CHRNA6 and CHRNB3) are associated with subjective responses to tobacco. Hum Mol Genet. 2008 Mar 1;17(5):724–34. doi: 10.1093/hmg/ddm344. [DOI] [PubMed] [Google Scholar]
- 21.Johnson EO, Morgan-Lopez AA, Breslau N, Hatsukami DK, Bierut LJ. Test of measurement invariance of the FTND across demographic groups: assessment, effect size, and prediction of cessation. Drug Alcohol Depend. 2008 Mar 1;93(3):260–70. doi: 10.1016/j.drugalcdep.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 22.Foroud T, Edenberg HJ, Goate A, Rice J, Flury L, Koller DL, et al. Alcoholism susceptibility loci: confirmation studies in a replicate sample and further mapping. Alcohol Clin Exp Res. 2000 Jul;24(7):933–45. [PubMed] [Google Scholar]
- 23.Laurie CC, Doheny KF, Mirel DB, Pugh EW, Bierut LJ, Bhangale T, et al. Quality Control and Quality Assurance in Genotypic Data for Genome-Wide Association Studies. Genetic Epidemioogy. doi: 10.1002/gepi.20516. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006 Aug;38(8):904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 25.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007 Sep;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanna S, Jackson AU, Nagaraja R, Willer CJ, Chen WM, Bonnycastle LL, et al. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat Genet. 2008 Feb;40(2):198–203. doi: 10.1038/ng.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008 Feb;40(2):161–9. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saccone NL, Wang JC, Breslau N, Johnson EO, Hatsukami D, Saccone SF, et al. The CHRNA5-CHRNA3-CHRNB4 nicotinic receptor subunit gene cluster affects risk for nicotine dependence in African-Americans and in European-Americans. Cancer Res. 2009 Sep 1;69(17):6848–56. doi: 10.1158/0008-5472.CAN-09-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ge D, Zhang K, Need AC, Martin O, Fellay J, Urban TJ, et al. WGAViewer: software for genomic annotation of whole genome association studies. Genome Res. 2008 Apr;18(4):640–3. doi: 10.1101/gr.071571.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spitz MR, Amos CI, Dong Q, Lin J, Wu X. The CHRNA5-A3 region on chromosome 15q24–25.1 is a risk factor both for nicotine dependence and for lung cancer. J Natl Cancer Inst. 2008 Nov 5;100(21):1552–6. doi: 10.1093/jnci/djn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ioannidis JP, Ntzani EE, Trikalinos TA. ‘Racial’ differences in genetic effects for complex diseases. Nat Genet. 2004 Dec;36(12):1312–8. doi: 10.1038/ng1474. [DOI] [PubMed] [Google Scholar]
- 32.Richardson CG, Ratner PA. A confirmatory factor analysis of the Fagerstrom Test for Nicotine Dependence. Addict Behav. 2005 May;30(4):697–709. doi: 10.1016/j.addbeh.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 33.Landgren S, Engel JA, Andersson ME, Gonzalez-Quintela A, Campos J, Nilsson S, et al. Association of nAChR gene haplotypes with heavy alcohol use and body mass. Brain Res [Research Support, Non-US Gov’t] 2009 Dec 11;1305( Suppl):S72–9. doi: 10.1016/j.brainres.2009.08.026. [DOI] [PubMed] [Google Scholar]
- 34.Hoft NR, Corley RP, McQueen MB, Huizinga D, Menard S, Ehringer MA. SNPs in CHRNA6 and CHRNB3 are associated with alcohol consumption in a nationally representative sample. Genes Brain Behav [Research Support, NIH, Extramural] 2009 Aug;8(6):631–7. doi: 10.1111/j.1601-183X.2009.00495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.