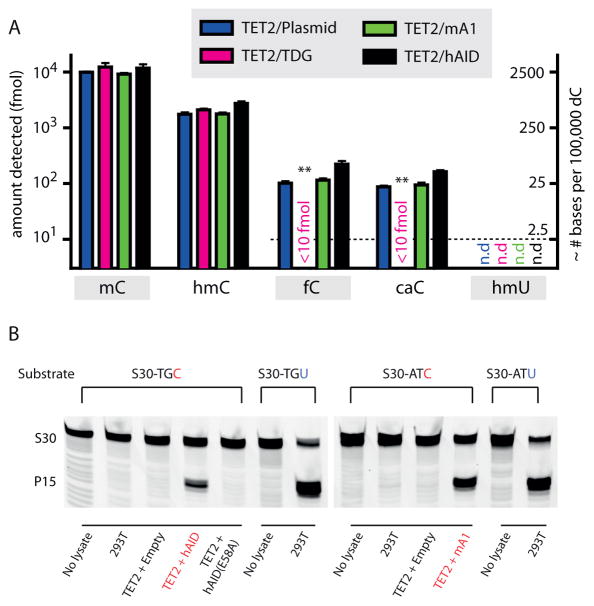
Figure 4. AID/APOBEC enzymes do not perturb levels of mC oxidation intermediates in genomic DNA.
(a) Genomic DNA was extracted from HEK 293T cells co-expressing TET2 along with an empty vector control, TDG, mA1 or hAID, digested to single nucleosides, and analyzed via mass spectrometry for the presence of C, mC, hmC, fC, caC, and hmU. The left axis depicts the absolute fmol of nucleoside on a logarithmic scale, while the right axis represents the approximate conversion from absolute fmol to the genomic prevalence of modified bases for every 105 dC bases. The dashed line demonstrates the lowest examined amount of fC, caC, and hmU standards. Error bars indicate the standard deviation from the mean for two to three biological replicates. Asterisks: p ≤ 10−3 for fC and caC in samples with TDG in comparison to plasmid only control. (b) To confirm that overexpressed hAID and mA1 are catalytically active, nuclear extracts were tested for deaminase activity against unmodified cytosine. At left, TET2-hAID nuclear extracts and negative controls (no extract, nuclear extract from untransfected 293T cells, and TET2-hAID E58A) were incubated with S30-TGC substrate; At right, TET2-mA1 nuclear extracts and negative controls (no extract and nuclear extract from untransfected 293T cells) were incubated with S30-ATC substrate. The lanes demonstrating deamination of S30-TGC by TET2-hAID and S30-ATC substrate by TET2-mA1 extracts are highlighted (red). Nuclear extracts were also incubated with S30-TGU or S30-ATU to verify robust uracil excision activity. Gels are shown without cropping in Supplementary Fig. 13.