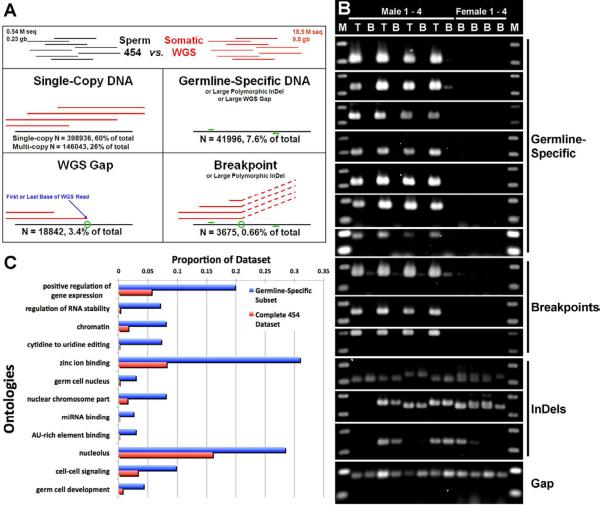
Figure 3. Analysis of pilot 454 sequencing data.
(A) All 454 reads were categorized on the basis of alignment patterns with the complete lamprey WGS dataset (liver DNA). A majority (82%) of reads appeared as “normal” DNA (multicopy or single-copy). Other alignment patterns were consistent with coverage gaps in the WGS dataset (3.4%), germline-specific DNA (7.6%) or recombination breakpoints (0.66%). Green circles depict the positions of alignment breaks and green arrows depict the generic locations of primer binding sites for validation PCRs. (B) Results of PCR validations of germline-specific/gene-containing (BLAST hit) reads and breakpoint-flanking reads provided positive validation of members of both rearrangement classes and identified segregating (in the population) insertion/deletion (InDel) polymorphisms and WGS coverage gaps, which mimic programmed rearrangement outcomes. Note, the “Germline-Specific” and “Breakpoint” classes result in similar PCR validation patterns because one primer (breakpoint) or both primers (germline-specific) are designed to germline-specific regions. T – template is testes DNA, B – template is blood DNA, M = 100 bp DNA ladder. (C) Overrepresented gene ontologies from 234 predicted germline-specific genes, relative to the entire 454 dataset (p>1e-8, corrected using false discovery rate control, as implemented by Blast2Go [9]).