Abstract
Biological invasions with known histories are rare, especially in the sea, and empirical studies of the genetic consequences are even rarer. Fifty-five years ago, the state of Hawai‘i began a remarkable, if unintentional, ‘experiment’ with the introduction of three reef fishes, Lutjanus fulvus, Cephalopholis argus and Lutjanus kasmira. All have since expanded from the initial introduction of 2204 to 3163 individuals; however, historical records show that initially L. fulvus remained scarce, C. argus had modest population expansion and L. kasmira experienced rapid population growth. The consequences of differential population growth rates are apparent in F-statistics: Hawaiian L. fulvus demonstrate strong and significant haplotype frequency shifts from the founder location (FST = 0.449), C. argus shows low but significant differentiation (FST = 0.066) and L. kasmira is nearly identical to the founder location (FST = 0.008). All three species had higher mtDNA diversity in the introduced range, which can be explained by multiple sources for L. fulvus and L. kasmira, but not for C. argus. We conclude that lag time before population expansion, in conjunction with genetic drift, has defined the genetic architecture of these three species in the introduced range.
Keywords: alien species, invasive, bottleneck, genetic drift, mtDNA, Papahānaumokuākea
1. Introduction
The dramatic decrease in effective population size (Ne) that accompanies founder events is expected to lead to decreased genetic diversity [1,2], and perhaps inbreeding depression and reduced evolutionary potential [3]. Genetic drift, which purges genetic diversity, exerts its greatest effects on founder populations during the early stages when population sizes are small [4]. Temporal and spatial delays in proliferation following a founder event, known as lag periods [5], can have substantial effects on genetic architecture [6] and have been attributed to: (i) Allee effects where low population densities result in decreased reproductive success [7–10], (ii) evolutionary factors, such as the time required for recombination and adaptation to novel environments [11,12], and (iii) ecological limitations such as the absence of facilitative mutualists [13].
Loss of genetic diversity following an introduction is not an inevitable outcome ([14–17], reviewed in [2]). High genetic diversity in the introduced range can be maintained if propagule pressure (density at the introduction site) is high or multiple introduction events take place [18,19]. In some cases, the introduction of individuals from genetically divergent populations can lead to higher genetic diversity in the introduced range compared with any single natural population [20–22]. Rapid population expansion early in an introduction decreases the likelihood of reduced diversity by genetic drift [2]. However, rarely are the details of introduction events known, so while there is a theoretical understanding of the impacts of propagule pressure, rate of population expansion and genetic drift on the genetic diversity of introduced populations, empirical data are scarce. Consequently, intentional and well-documented introductions are invaluable case studies for evaluating the effects of founder population size and the impact of lag periods and stochastic lineage sorting on patterns of genetic diversity.
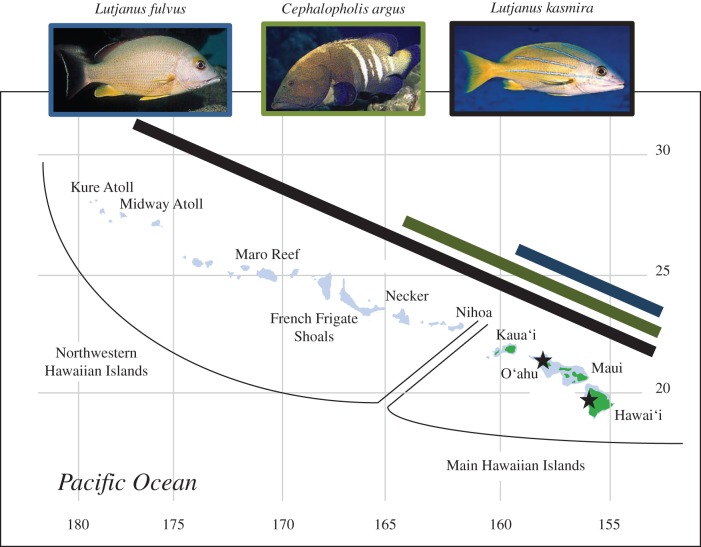
Half a century ago, the Hawai‘i Division of Fish and Game (HDFG, now Hawai‘i Division of Aquatic Resources (DAR); http://hawaii.gov/dlnr/dar/index.html) undertook an ambitious fishery-enhancement programme by introducing 12 species of snappers and groupers to the Hawaiian Islands [23–25]. Three became established: the blacktail snapper, Lutjanus fulvus, the peacock hind, Cephalopholis argus, and the bluestriped snapper, Lutjanus kasmira. The introductions occurred in several events between 1955 and 1961 (see the electronic supplementary material, table S1, HDFG records: DAR, Honolulu, HI, USA). Within 15 years, all three species had been recorded throughout the main Hawaiian Islands (MHI, figure 1): successes that may have been fostered by ecological release from competitors and parasites [26–28] in the depauperate Hawaiian reef ecosystems. However, HDFG records indicate that population densities of L. fulvus remained low following introduction, C. argus was more common, while L. kasmira proliferated rapidly, reaching Midway Atoll in the far northwest of the archipelago (greater than 2100 km from the introduction point) by 1992 [23]. Combining genetic analyses with HDFG records shows that the rapid colonization of the archipelago by L. kasmira was accompanied by maintenance of high levels of genetic diversity, indicating large numbers of colonists at every island along the way [29]. In contrast to the highly successful L. kasmira, C. argus has spread only halfway up the archipelago to French Frigate Shoals (FFS), with a total range of approximately 1200 km, while L. fulvus has remained restricted to the MHI, with a total range of approximately 600 km (figure 1) [24,30]. While the introductions were well intentioned, these species have not become popular food fishes in Hawai‘i and are now largely viewed as a threat to native Hawaiian fauna.
Figure 1.
Map of the Hawaiian archipelago showing sample locations and the introduced range of L. fulvus (blue bar), C. argus (green bar) and L. kasmira (black bar). Black stars indicate sites of introduction (see electronic supplementary material, table S1 for details). Photo credit: Keoki and Yuko Stender.
The well-documented introduction of L. fulvus, C. argus and L. kasmira to the Hawaiian Islands in roughly equal numbers (see the electronic supplementary material, table S1) provides a rare opportunity to directly evaluate the impact of lag period on genetic diversity and invader success. Here we combine historical records with mitochondrial cytochrome b (cyt b) sequence data from L. fulvus, C. argus and L. kasmira to investigate their introduction to the Hawaiian Islands and to ask the following questions: (i) Is there evidence of a loss of genetic diversity at the introduction site or at more distant Hawaiian islands, as expected under a stepping-stone series of colonizations? (ii) Did the differing rates of population growth impact genetic diversity? (iii) Can the differential spread of these three species in Hawai‘i be explained by differences in patterns of genetic diversity; or (iv) Are other proximate causes responsible for the differential success of these three species? Here we capitalize on these well-documented introductions to Hawai‘i to examine the impact of lag period on genetic architecture in a comparative framework.
2. Material and methods
(a). Historical records
In 1955, the Territory of Hawai‘i instigated Project no. F-5-R ‘The introduction of marine game fishes from areas in the Pacific’ to introduce desirable shallow-water game and food fishes from the tropical and subtropical Pacific. Progress reports were filed and included surveys of native habitats for suitable species, details of the introductions, underwater observations of introduced species by Fish and Game officers and sightings reported by local fishermen. While quantitative fish counts are not included in these reports, fish sightings and in most cases the numbers of fishes were recorded. Successive sightings on a new island was considered a range expansion. While colonization of new islands is not necessarily coupled with population growth, it is evidence of reproduction (dispersal in these fishes occurs largely during the larval phase and movement of adults across open channels has not been documented).
(b). Study species and collections
The blacktail snapper, L. fulvus (Schneider 1801), the peacock hind, C. argus (Bloch and Schneider 1801) and the bluestriped snapper, L. kasmira (Forsskål 1775), occupy nearly the same geographical range, from the Marquesas Islands in the central Pacific to the east coast of Africa. Previously, genetic diversity was characterized across the natural range of all three species [31,32]. A total of 157 new specimens of L. fulvus were collected from four locations, and 236 specimens of C. argus from seven locations across the Hawaiian archipelago by scuba divers using polespears (table 1 and figure 1). A subset of the L. kasmira specimens from Gaither et al. [29] (O‘ahu = 44; Hawai‘i Island = 49) were used in this study (table 1). Specimens from the uninhabited northwestern Hawaiian Islands were obtained during research expeditions on the NOAA R/V Hi‘ialakai, as part of an initiative to aid the Papahānaumokuākea Marine National Monument (http://hawaiireef.noaa.gov/) in efforts to monitor and characterize this vast protected area. Tissue samples (fin clips or gill filaments) were preserved in either 95 per cent ethanol (EtOH) or saturated NaCl solution [33,34], and stored at room temperature.
Table 1.
Molecular diversity indices for mitochondrial cytochrome b (cyt b) sequences. Lutjanus fulvus and C. argus = cyt b (data for source populations from Gaither et al. [31,32], respectively), L. kasmira = control region (data from Gaither et al. [29]); Hawai‘i Island and Maui in the current table = Kona + Hilo and Maui Nui in original dataset, respectively); data in parentheses are cyt b data generated from a subset (N = 189 of 484) of the samples from Gaither et al. [29]. Number of specimens (n), number of haplotypes (nh), haplotype diversity (h) and nucleotide diversity (π) as reported by Arlequin v. 3.5 [36] are listed. FFS, French Frigate Shoals. Source locations = Nuku Hiva, Marquesas Islands; Moorea, Society Islands; Kanton, Phoenix Islands.
|
L. fulvus |
C. argus |
L. kasmira |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cyt b |
cyt b |
control region (cyt b) |
||||||||||
| N | Nh | h | π | N | Nh | h | π | N | Nh | h | π | |
| source location | ||||||||||||
| Marquesas | 48 | 7 | 0.72 ± 0.04 | 0.002 ± 0.002 | — | — | — | — | 50 (47) | 47 (18) | 1.00 ± 0.01 (0.83 ± 0.04) | 0.019 ± 0.010 (0.004 ± 0.003) |
| Society | 48 | 4 | 0.12 ± 0.06 | 0.001 ± 0.001 | 44 | 6 | 0.39 ± 0.09 | 0.001 ± 0.001 | 49 (49) | 31 (10) | 0.97 ± 0.01 (0.51 ± 0.09) | 0.016 ± 0.009 (0.002 ± 0.001) |
| Phoenix | 46 | 10 | 0.66 ± 0.07 | 0.005 ± 0.003 | — | — | — | — | — | — | — | — |
| all source | 142 | 17 | 0.70 ± 0.03 | 0.006 ± 0.004 | 44 | 6 | 0.39 ± 0.09 | 0.001 ± 0.001 | 99 (96) | 78 (28) | 0.99 ± 0.01 (0.83 ± 0.03) | 0.038 ± 0.019 (0.005 ± 0.003) |
| introduced range | ||||||||||||
| Hawai‘i Island | 40 | 7 | 0.73 ± 0.04 | 0.007 ± 0.004 | 51 | 5 | 0.52 ± 0.07 | 0.001 ± 0.001 | 101 (49) | 41 (19) | 0.99 ± 0.01 (0.89 ± 0.03) | 0.038 ± 0.019 (0.005 ± 0.003) |
| Maui | 41 | 6 | 0.75 ± 0.04 | 0.007 ± 0.004 | 53 | 4 | 0.55 ± 0.04 | 0.001 ± 0.001 | 39 | 32 | 0.99 ± 0.01 | 0.034 ± 0.017 |
| O‘ahu | 40 | 7 | 0.73 ± 0.05 | 0.007 ± 0.004 | 53 | 6 | 0.61 ± 0.05 | 0.001 ± 0.001 | 50 (44) | 40 (19) | 0.99 ± 0.01 (0.93 ± 0.02) | 0.033 ± 0.017 (0.005 ± 0.003) |
| Kaua‘i | 36 | 7 | 0.77 ± 0.05 | 0.007 ± 0.004 | 53 | 7 | 0.59 ± 0.05 | 0.001 ± 0.001 | 36 | 30 | 0.99 ± 0.01 | 0.035 ± 0.018 |
| Nihoa | — | — | — | — | 13 | 3 | 0.56 ± 0.11 | 0.001 ± 0.001 | — | — | — | — |
| Necker | — | — | — | — | 9 | 3 | 0.56 ± 0.17 | 0.001 ± 0.001 | 49 | 38 | 0.99 ± 0.01 | 0.035 ± 0.018 |
| FFS | — | — | — | — | 4 | 2 | 0.50 ± 0.27 | 0.001 ± 0.001 | 40 | 38 | 1.00 ± 0.01 | 0.033 ± 0.017 |
| Maro | — | — | — | — | — | — | — | — | 21 | 18 | 0.98 ± 0.02 | 0.036 ± 0.018 |
| Midway | — | — | — | — | — | — | — | — | 40 | 33 | 0.99 ± 0.01 | 0.046 ± 0.023 |
| Kure | — | — | — | — | — | — | — | — | 9 | 9 | 1.00 ± 0.05 | 0.034 ± 0.019 |
| all Hawai‘i | 157 | 8 | 0.74 ± 0.02 | 0.007 ± 0.004 | 236 | 10 | 0.57 ± 0.24 | 0.001 ± 0.001 | 385 (93) | 172 (27) | 0.99 ± 0.01 (0.91 ± 0.02) | 0.037 ± 0.018 (0.006 ± 0.003) |
(c). DNA extraction, PCR amplifications and sequencing
Mitochondrial cyt b sequences were obtained using protocols (DNA extraction, PCR cycling and sequencing) identical to those described by Gaither et al. [31] for L. fulvus and L. kasmira and Gaither et al. [32] for C. argus. Cyt b sequences were obtained from all source populations (L. fulvus: Marquesas, Society and Phoenix Islands; C. argus: Society; L. kasmira: Marquesas and Society; N = 44–50, table 1). Sequences for each species were aligned and edited using Geneious Pro v. 5.0 (Biomatters Ltd., Auckland, New Zealand). In all cases, alignment was unambiguous with no indels or frameshift mutations. Haplotypes used in this study were labelled (see the electronic supplementary material, table S2) and deposited in GenBank (accession numbers: L. fulvus: JX316840–JX316858; C. argus: JX316859–JX316869; L. kasmira: JX316870–JX316910).
(d). Data analysis
Summary statistics, including haplotype diversity (h) and nucleotide diversity (π), were estimated with algorithms from Nei [35] as implemented in Arlequin v. 3.5 [36]. Phylogenetic median-joining networks were constructed using Network v. 4.5 with default settings [37]. Analyses of molecular variance (AMOVAs) were performed in Arlequin using 20 000 permutations. Wright's FST was calculated to detect significant haplotype frequency shifts and was not used to measure conventional population structure or to make estimates of migration.
To compare genetic diversity in the introduced and source populations, we estimated haplotype frequencies in a founder population by weighting the relative contribution of each source population, based on contemporary cyt b diversity. To control for unequal sample sizes [38], we estimated haplotype richness using rarefaction analysis (Analytic Rarefactation v. 1.4; UGA Stratigraphy Lab website; http://www.uga.edu/~strata/software/). Owing to the lower sensitivity of heterozygosity to losses of genetic diversity [1], we examined changes in haplotype richness to estimate the impact of the founding event on genetic diversity. We used a χ2-test (GraphPad Software, http://www.graphpad.com/welcome.htm) to determine whether the number of unique haplotypes (found only in either the native or introduced range) differed significantly from the null expectation that unique haplotypes were evenly distributed among regions.
3. Results
For each species, sample size (N), number of haplotypes (Nh), h and π per location are listed in table 1.
(a). Lutjanus fulvus
We resolved a 480 bp segment of cyt b in 157 Hawaiian individuals and analysed these with 142 sequences from source populations [31]. We detected 19 haplotypes: 17 in the source populations and eight in Hawai‘i (table 1 and figure 2a). Among the source populations, π = 0.001–0.005, while the corresponding haplotype diversity indices were h = 0.12–0.72 (table 1). Hawaiian samples demonstrated significantly higher values of π (0.007, Welch one-tailed t-test, t = 8.66, d.f. = 2, p = 0.001) and consistently (although not statistically significant) higher values of h (range = 0.73–0.77, Welch one-tailed t-test, t = 1.28, d.f. = 2, p = 0.164) compared with the source populations. There was a highly significant shift in haplotype frequencies between the founder population and the introduced population in Hawai‘i (figure 3a; FST = 0.449, p < 0.001; Fisher's exact test, p < 0.001).
Figure 2.
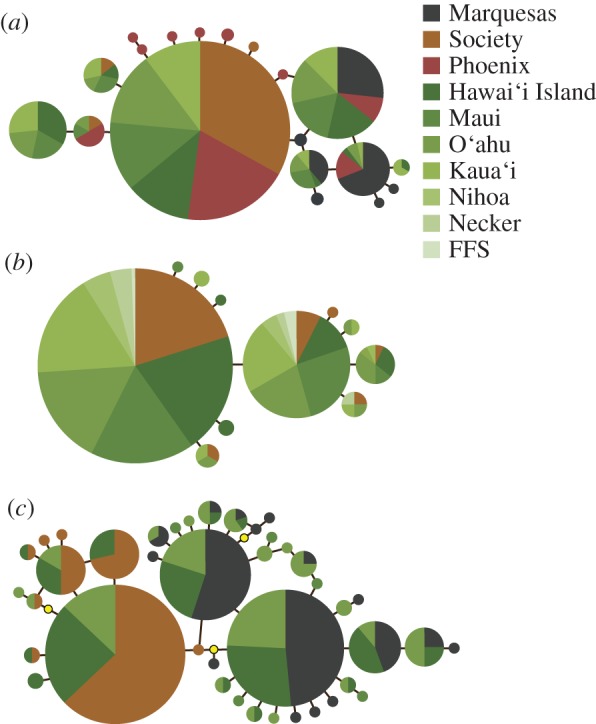
Median-joining networks for cytochrome b sequences from (a) L. fulvus, (b) C. argus, and (c) L. kasmira constructed using default settings in the program Network v. 4.5 [37]. Lutjanus kasmira cyt b sequences generated from a subset (N = 189 of 484) of the samples from Gaither et al. [29]. Each circle represents one mitochondrial haplotype with the area of each circle proportional to the number of that particular haplotype in the dataset; yellow circles represent missing haplotypes; colours represent sampling location (see key). FFS = French Frigate Shoals. Source locations = Nuku Hiva, Marquesas Islands; Moorea, Society Islands; Kanton, Phoenix Islands.
Figure 3.
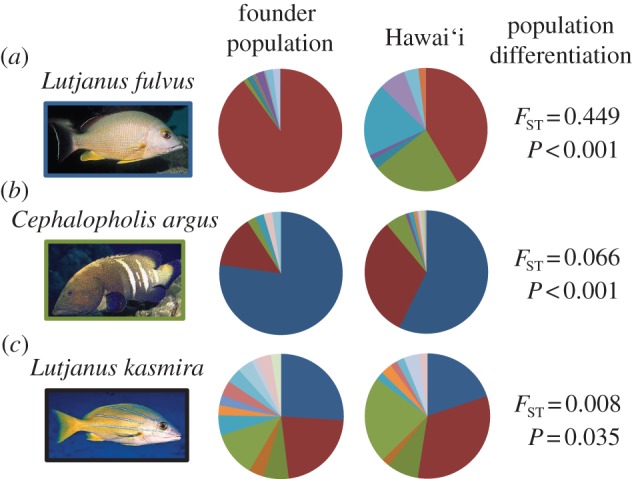
Haplotype frequencies for (a) L. fulvus, (b) C. argus, and (c) L. kasmira. We estimated cyt b haplotype distributions in founder populations as the product of contemporary haplotype frequencies in the source populations and the number of individuals introduced from each of those sources (see the electronic supplementary material, tables S1 and S2). Differences in haplotype frequencies between the founder and introduced populations were detected using AMOVA and an exact test of population differentiation as implemented in Arlequin v. 3.5 [36]. The resulting F-statistic and the associated p-value from the AMOVAs are shown. All comparisons were significant using the exact test (p < 0.001; data not shown). Photo credit: Keoki and Yuko Stender.
Lutjanus fulvus haplotypes are closely related, differing by only 1–6 bp (figure 2a). Two of the most common haplotypes in the native range were detected at each sample location in Hawai‘i (see the electronic supplementary material, table S2). Putative private haplotypes were found in each source population (see the electronic supplementary material, table S2; Marquesas = Lfu12–Lfu16; Society = Lfu17, Lfu19; Phoenix = Lfu1, Lfu4, Lfu6–Lfu9). One private haplotype from the Marquesas (Lfu16) and one from the Society population (Lfu17) were detected in the introduced range. None of the six private haplotypes in the Phoenix population were detected in Hawai‘i. Two of the eight haplotypes in Hawai‘i went undetected in the native range; one of which accounts for nearly 20 per cent of the individuals in the introduced range (see the electronic supplementary material, table S2; Lfu11).
The presence of the putative Marquesan haplotype Lfu16 at all MHI locations (see the electronic supplementary material, table S2) indicates that despite the low number of individuals introduced from this location (N = 35), the descendants of these individuals spread throughout the introduced range. As expected for introductions from multiple sources, the Hawaiian samples had a greater number of haplotypes (Nh = 8) and higher diversity indices (h = 0.73–0.77) than the Society Islands’ population, which accounted for 92 per cent of the founders (table 1).
When the dataset was grouped by native versus introduced ranges, we found significant overall structure in the native range (Marquesas, Society and Phoenix Islands) for L. fulvus (FST = 0.705, p < 0.001) with particular distinction of the Marquesas population [31]. We found no significant haplotype frequency shifts among the introduced locations (FST = −0.021, p = 0.992). The Marquesas and Society (source) populations were significantly different from each of the four Hawaiian samples with highest levels of structure between the Marquesas and introduced populations (FST = 0.523–0.549) and lower but significant values between Society and Hawaiian populations (FST = 0.280–0.291). The Phoenix population was not significantly different from any of the introduced populations (see the electronic supplementary material, table S3).
(b). Cephalopholis argus
We resolved a 615 bp segment of cyt b in 236 individuals from Hawai‘i and analysed these with 44 sequences from the source population at the Society Islands [32]. We detected 11 haplotypes: six in the source population and 10 in the introduced range (table 1 and figure 2b). Nucleotide diversity was low and did not vary among samples (π = 0.001). Haplotype diversity was low in the source population (h = 0.39) and, contrary to expectation, significantly higher in all Hawaiian samples (range = 0.50–0.61; one sample t-test, t = 11.60, d.f. = 6, p < 0.001). Cephalopholis argus demonstrated two dominant haplotypes both of which were detected in the native and introduced ranges (see the electronic supplementary material, table S2 and figure 2b). Similar to L. fulvus, there is a significant, although less dramatic, shift in allele frequencies between the source population and Hawai‘i (figure 3b; FST = 0.066, p < 0.001; Fisher's exact test, p < 0.001). The shift in haplotype frequencies, together with the detection of five putative private haplotypes in Hawai‘i, accounts for the higher genetic diversity in the introduced range (table 1).
When the dataset is grouped by native versus introduced ranges, we found that three of the seven samples in Hawai‘i (Maui, O‘ahu and FFS) differed significantly from the source population (FST = 0.070–0.309, electronic supplementary material, table S4) with no significant haplotype frequency shifts among the samples in the introduced range (see the electronic supplementary material, table S4).
(c). Lutjanus kasmira
We resolved a 447 bp segment of cyt b in a subset of the L. kasmira specimens (O‘ahu = 44; Hawai‘i Island = 49; table 1) from Gaither et al. [29] and combined these with 96 cyt b sequences from the source populations [31]. We detected 41 haplotypes: 28 in the source populations and 27 in the introduced range. As expected from a population of mixed lineages, Hawaiian samples demonstrated consistently (although not statistically significant) higher values of h and π compared with the source populations. When we reconstructed the genetic composition of the founder population, there was not a considerable haplotype frequency shift following the introduction of L. kasmira to Hawai‘i (figure 3c; FST = 0.008, p = 0.035; Fisher's exact test, p < 0.001), as observed in L. fulvus and, to a lesser extent, in C. argus.
(d). Genetic diversity
We observed 17 cyt b haplotypes in L. fulvus in the native range (N = 142) (table 1), but only eight haplotypes in the slightly larger sample from Hawai‘i (N = 157). Of the 17 haplotypes recorded in the native population, 11 were putative private haplotypes compared with only two in Hawai‘i. After correcting for sample size, the difference in the number of private haplotypes observed in the native and introduced populations was significant (χ2 = 7.181, d.f. = 1, p = 0.007); however, no such difference was detected in either C. argus (χ2 = 0.002, d.f. = 1, p = 0.965) or L. kasmira (χ2 = 0.008, d.f. = 1, p = 0.929).
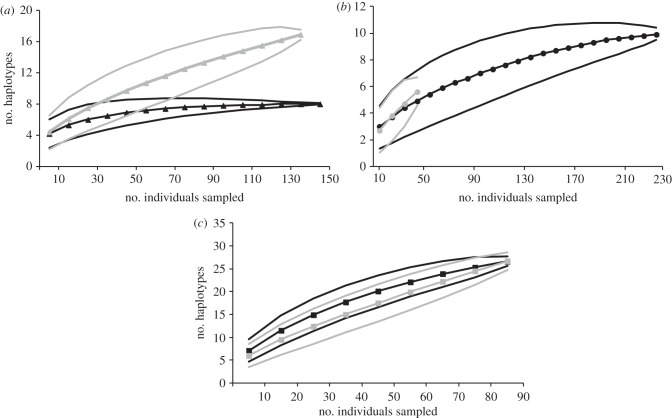
Owing to the large confidence intervals (95%) of the rarefaction curves, there was no significant difference between the expected number of mtDNA haplotypes in the native and introduced ranges at low sample sizes (figure 4). However, as sample size increased (N > 50), the curves for L. fulvus no longer overlap (figure 4a) and a loss of mtDNA haplotypes in the introduced range became evident in this species. No loss of haplotypes was detected in either C. argus or L. kasmira using rarefaction (figure 4b,c).
Figure 4.
Rarefaction curves plotting the number of individuals sampled against the expected number of mitochondrial haplotypes were calculated using the Analytic Rarefactation v. 1.4 software available at the UGA Stratigraphy Lab website (http://www.uga.edu/~strata/software/) for (a) L. fulvus, (b) C. argus, and (c) L. kasmira. Grey lines represent data from the source populations and black lines represent data from the introduced range. Also, 95% confidence limits are shown (number of species ±1.96 * sqrt (var)).
4. Discussion
Our surveys of introduced fishes in Hawai‘i provide a rare opportunity to examine the empirical relationship between lag period and patterns of genetic diversity after founder events. We found maintenance of high levels of diversity and little to no change in haplotype frequencies (FST = 0.008) in the rapidly expanding L. kasmira, high diversity and a small (but significant) shift in haplotype frequencies (FST = 0.066) in the moderately expanding C. argus and a contrasting loss of diversity and drastic shift in haplotype frequencies in the slowly expanding L. fulvus (FST = 0.449; figure 3). In the introduced range, we detected only eight haplotypes in L. fulvus compared with 17 in the source populations. In contrast, only one haplotype detected in the native range of C. argus went undetected in Hawai‘i, an introduced population that harboured five haplotypes not detected in the native range. These findings indicate that the observed genetic architectures were defined by the pace of population expansion in the introduced range, and presumed population growth.
Prior to dissecting these results, we discuss one primary caveat: the analyses presented here assume that haplotype frequencies in the contemporary source locations are a suitable surrogate for the haplotype composition of the original source populations (55 years ago).
(i) Did we sample the same source populations? We sampled populations at the island from which the original founders were derived. The larval duration of these species are estimated at between 30 and 50 days, making fine-scale genetic structure among sites at a single island unlikely. Further, range-wide genetic surveys for these species [31,32] indicate genetic connectivity on the scale of ocean basins (tens of thousands of kilometres) for L. kasmira and C. argus, and over thousands of kilometres for L. fulvus.
(ii) Could haplotype frequencies have changed in the source populations over the last 55 years? Assuming neutrality, changes in allele frequencies are determined by genetic drift and mutation. The average mutation rate for cyt b is estimated at roughly 1 per cent per million years [39–41], indicating that mutations in this gene fragment over the past 55 years should be negligible. The impact of drift accumulates over generations, and is proportional to effective population size. Using a generation time of 5 years for these species [31,32], we estimate that at most 12 generations have passed since the introductions took place: a short time for genetic drift to act, especially if the population sizes are large. However, if population sizes in the native range were very small, then genetic drift could have a significant influence in only a few generations. This is not the case for the primary source locations (French Polynesian), with first-order estimates of contemporary female effective population size (Nef based on mtDNA) of 100 000 for C. argus and an order of magnitude higher for each of the snappers [31,32]. Because population sizes of these three species are large, and time since introduction is short, we regard the recent samples from source locations as a reasonable proxy for the original source populations.
(a). Establishment and spread in Hawai‘i
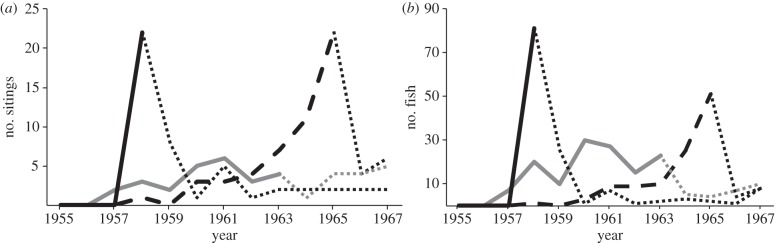
The introduction of L. fulvus, C. argus and L. kasmira to Hawai‘i occurred in several events between 1955 and 1961 (see the electronic supplementary material, table S1, HDFG records). Within 15 years, all three species had been recorded throughout the MHI (figure 1). While quantitative surveys were not conducted following the introductions, catch reports by fishermen and observations by HDFG officers reveal compelling patterns. HDFG records indicate that population densities of L. fulvus remained low following introduction, with a gradual increase in sightings that peak in 1965 at 22 events recording 51 fish (figure 5). Cephalopholis argus followed a similar pattern of establishment but a faster rate of spread than L. fulvus, with HDFG records indicating that population sizes grew steadily after the introduction (figure 5). In comparison, L. kasmira was a rapid and prolific invader that quickly spread through the archipelago at a rate of about 60 km per year [24,30]. In 1992, just 34 years after the initial introduction, L. kasmira was recorded at the far reaches of the archipelago at Midway Atoll [42] over 2100 km from the release site. HDFG catch records indicate that population densities remained high after the introduction. Lutjanus kasmira was first recorded in Hawaiian waters in 1958 when 22 sightings reported a total of 88 fish (figure 5). After that year, records were discontinued owing to the commonality of large schools (more than 50 individuals). In 1970, commercial fishermen reported landing 0.5 metric tonne of L. kasmira. By 1981, this number had grown to 37 metric tonnes; a level of exploitation far exceeding the other two species [43].
Figure 5.
Data from State of Hawai‘i Division of Fish and Game (HDFG) records for 12 years following the introductions. (a) The number of sightings and (b) the total number of fishes caught by local fishers or seen by HDFG employees during surveys are recorded for L. fulvus (dashed black line), C. argus (grey line) and L. kasmira (solid black line). Dotted lines represent the point at which records were no longer kept and/or reports by local fishers decreased owing to commonality of sightings.
The cause of the inferred differential rates of population growth in these species is unknown. Lag periods, such as those demonstrated by L. fulvus in Hawai‘i, occur when there are relatively slow rates of population growth or range expansion following introduction and are often attributed to a variety of factors, including low population densities (Allee effect) or environmental and ecological impediments to expansion (reviewed in [5]). Considered in isolation, the slower growth of L. fulvus may not have been interpreted as a lag time, but in this comparative framework, population density of this species remained low for an extended period relative to the other two species. This pattern could simply reflect differences in intrinsic growth rates among the three species; however, there are insufficient data to test this hypothesis.
(b). Genetic effects of the founder events
The lack of genetic structure across the introduced range of these fishes, coupled with the maintenance of genetic diversity, contradicts a stepping-stone model of colonization (reviewed in [44]). Genetic surveys across 27 taxa, including invertebrates, fish and mammals, in Hawai‘i identify barriers to dispersal at Kaua‘i and Hawai‘i Island in more than half the species surveyed [45]. We did not find evidence for barriers to dispersal within the MHI in these fishes. Our data, in conjunction with HDFG records, indicate that these species colonized each island in sufficient numbers to capture most of the standing genetic diversity, or that subsequent gene flow was sufficient to homogenize the geographical distribution of the genetic diversity, or both.
Following the introduction of L. fulvus, there was a highly significant shift in haplotype frequencies. The majority of L. fulvus released in Hawai‘i (92%) were derived from the Society Islands (see the electronic supplementary material, table S1). This population is nearly homogenous for haplotype Lfu2 (see the electronic supplementary material, table S2), yet this haplotype constitutes just 41 per cent of the genetic diversity in the introduced range. Only eight haplotypes were detected in Hawai‘i compared with 15 in the native range. The missing haplotypes were rare in the native range and their loss in Hawai‘i is evident from the rarefaction analysis (figure 4). One haplotype in Hawai‘i (Lfu11) was not detected in the source populations, yet represents nearly 20 per cent of individuals sampled in the introduced range. Together, these findings point to stochastic lineage sorting during the early stages of the introduction when population densities of L. fulvus were low, and variability in reproductive success was probably high.
Contrary to expectations, we found higher haplotype diversity in the Hawaiian populations of C. argus than in the source population. The Society Islands population of C. argus, similar to L. fulvus, is dominated by a single haplotype (Car1 at 77%, electronic supplementary material, table S2). However, this haplotype constitutes just 57 per cent of the individuals in Hawai‘i while other haplotypes are found in greater frequency compared with the source population (Car2: 33% versus 14%; Car3 6% versus 2%, respectively). Five putative private haplotypes in Hawai‘i contribute to higher genetic diversity. While high diversity in introduced L. kasmira can be explained by multiple source populations, the higher diversity in C. argus is probably due to lower sampling effort at the source population (relative to the introduced population) in conjunction with stochastic shifts in haplotype frequencies following introduction (table 1 and electronic supplementary material, table S2b).
(c). The failed introductions
Nine other species of snappers and groupers were introduced during the same time period but failed to establish (see the electronic supplementary material, table S1). No phylogeographic data have been published for these species and few specific life-history data are available, leaving little information upon which to base hypotheses concerning their extirpation in Hawai‘i. Four species were introduced in low numbers (less than 200 individuals). One of these, L. gibbus, was spotted for several years following the introduction and thought to be the fourth successful introduction; however, no sightings have been recorded since the 1970s. Four other species were introduced in numbers greater than 900 individuals (see the electronic supplementary material, table S1). For instance, over 3400 L. guttatus were introduced to Hawai‘i from Mexico (the only species introduced from outside French Polynesia). There is no evidence that this species reproduced in Hawai‘i and catch records indicate a steady decline in numbers following the introduction. It is likely that this Eastern Pacific species, which primarily inhabits continental shelves, was maladapted to an oceanic island environment, but in none of these cases can stochasticity be ruled out.
5. Conclusions
Empirical studies that document the genetic consequences of marine invasions with known histories are essentially unknown. Here, we substantiate the importance of stochastic lineage sorting in shaping genetic architecture during the invasion process. Further, we show that the genetic architecture of founder populations can be significantly altered following introduction, making identification of source populations in undocumented invasions complicated. Lutjanus kasmira, which maintained high population densities and spread quickly, was able to retain much of the genetic diversity inherent in the native populations. In contrast, L. fulvus, which was slower ‘coming out of the starting blocks’, had substantial changes in genetic architecture, including significant loss of genetic diversity. Whether these losses influence the ultimate success of the species in Hawai‘i is unknown, but the loss of genotypes implies a loss of evolutionary potential. The finding of an intermediate shift in haplotype frequencies in C. argus, which maintained moderate population densities during the early stages of the introduction, implies that the changes we recorded are not due to directional selection in the new environment. The nature of this case study precludes replication, and therefore, we cannot conclusively rule out chance as driving the observed patterns. However, the available evidence supports our conclusion that the genetic architecture in the introduced range has been shaped primarily by lag time and corresponding genetic drift. Our data highlight that patterns of genetic diversity are influenced not simply by propagule pressure and the genetic diversity in source populations but perhaps of equal importance are the population growth trajectories and stochastic processes when population sizes are in flux.
Acknowledgements
We thank Papahānaumokuākea Marine National Monument co-trustees NOAA Marine Sanctuaries, US Fish and Wildlife Service (USFWS) and the State of Hawai‘i. This study was supported by the National Science Foundation (grants nos OIA0554657, OCE-0453167 and OCE-0929031 to B.W.B.), NOAA National Centers for Coastal Ocean Science Coral Reef Ecosystem Studies grant NA07NOS4780188, NOAA Coral Reef Conservation Programme and Hawai‘i Undersea Research Laboratory grant NA05OAR4301108, NOAA National Marine Sanctuaries Programme MOA grant no. 2005-008/66882 (R.J.T.) and NOAA Project R/HE-1, which is sponsored by the University of Hawai‘i Sea Grant College Programme, SOEST, under Institutional grant no. NA09OAR4170060. The views expressed herein are those of the authors and do not necessarily reflect the views of NSF, NOAA, USFWS, the State of Hawai‘i or any of their subagencies. Greg Concepcion, Matt Craig, Jonathan Dale, Toby Daly-Engel, Joseph DiBattista, Jeff Eble, Matt Iacchei, Shelley Jones, Stephen Karl, Randall Kosaki, Carl Meyer, Yannis Papastamatiou, Luiz Rocha, Zoltan Szabo, Jill Zamzow, Joshua Reece and the crew of the R. V. Hi‘ialakai helped collect specimens. Aulani Wilhelm, Jo-Ann Leong, Hoku Johnson, Danielle Carter, Daniel Polhemus, Randall Kosaki, Ann Mooney, Elizabeth Keenen and Kelly Gleason provided crucial logistic assistance to this project. We thank the members of the ToBo lab and staff at HIMB for their assistance and feedback and Molly Timmers who provided the map for figure 1. We especially thank the associate editor Daniel Ruzzante and four anonymous reviewers whose comments greatly improved this manuscript. This is contribution no. 1506 from the Hawai‘i Institute of Marine Biology, no. 8706 from the School of Ocean and Earth Science and Technology and no. UNIHI-SEAGRANT-JC-09-47 from the University of Hawai‘i Sea Grant Programme.
References
- 1.Nei M., Maruyama T., Chakraborty R. 1975. The bottleneck effect and genetic variability in populations. Evolution 29, 1–10 10.2307/2407137 (doi:10.2307/2407137) [DOI] [PubMed] [Google Scholar]
- 2.Dlugosch K. M., Parker I. M. 2008. Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol. Ecol. 1, 431–449 10.1111/j.1365-294X.2007.03538.x (doi:10.1111/j.1365-294X.2007.03538.x) [DOI] [PubMed] [Google Scholar]
- 3.Frankham R. 2005. Genetics and extinction. Biol. Conserv. 126, 131–141 10.1016/j.biocon.2005.05.002 (doi:10.1016/j.biocon.2005.05.002) [DOI] [Google Scholar]
- 4.Kimura M., Ohta T. 1969. The average number of generations until fixation of a mutant gene in a population. Genetics 61, 763–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crooks J. A. 2005. Lag times and exotic species: the ecology and management of biological invasions in slow-motion. Ecoscience 12, 316–329 10.2980/i1195-6860-12-3-316 (doi:10.2980/i1195-6860-12-3-316) [DOI] [Google Scholar]
- 6.Baker A. J., Moeed A. 1987. Rapid genetic differentiation and founder effect in colonizing populations of Common Mynas (Acridotheres tristis). Evolution 41, 525–538 10.2307/2409254 (doi:10.2307/2409254) [DOI] [PubMed] [Google Scholar]
- 7.Lewis M. A., Kareiva P. 1993. Allee dynamics and the spread of invading organisms. Theor. Pop. Biol. 43, 141–158 10.1006/tpbi.1993.1007 (doi:10.1006/tpbi.1993.1007) [DOI] [Google Scholar]
- 8.Drake J. M. 2004. Allee effects and the risk of biological invasion. Risk Analysis 24, 795–802 10.1111/j.0272-4332.2004.00479.x (doi:10.1111/j.0272-4332.2004.00479.x) [DOI] [PubMed] [Google Scholar]
- 9.Leung B., Drake J. M., Lodge D. M. 2004. Predicting invasions: propagule pressure and the gravity of Allee effects. Ecology 85, 1651–1660 10.1890/02-0571 (doi:10.1890/02-0571) [DOI] [Google Scholar]
- 10.Taylor C. M., Hastings A. 2005. Allee effects in biological invasions. Ecol. Lett. 8, 895–908 10.1111/j.1461-0248.2005.00787.x (doi:10.1111/j.1461-0248.2005.00787.x) [DOI] [Google Scholar]
- 11.Crooks J. A., Soulé M. E. 1999. Lag times in population explosions of invasive species: causes and implications. In Invasive species and biodiversity management (eds Sandlund O. T., Schei P. J., Viken A.), pp. 103–125 Boston, MA: Kluwer Academic Press [Google Scholar]
- 12.Lee C. E. 2002. Evolutionary genetics of invasive species. Trends Ecol. Evol. 17, 386–391 10.1016/S0169-5347(02)02554-5 (doi:10.1016/S0169-5347(02)02554-5) [DOI] [Google Scholar]
- 13.Richardson D. M., Allsopp N., D'Antonio C. M., Milton S. J., Rejmánek M. 2000. Plant invasions—the role of mutualisms. Biol. Rev. Camb. Phil. Soc. 75, 65–93 10.1111/j.1469-185X.1999.tb00041.x (doi:10.1111/j.1469-185X.1999.tb00041.x) [DOI] [PubMed] [Google Scholar]
- 14.Bossdorf O., Auge H., Lafuma L., Rogers W. E., Siemann E., Prati D. 2005. Phenotypic and genetic differentiation between native and introduced plant populations. Oecologia 144, 1–11 10.1007/s00442-005-0070-z (doi:10.1007/s00442-005-0070-z) [DOI] [PubMed] [Google Scholar]
- 15.Wares J. P., Hughes R., Grosberg K. 2005. Mechanisms that drive evolutionary change. In Species invasions: insights into ecology, evolution, and biogeography (eds Sax D. F., Stachowicz J. J., Gaines S. D.), pp. 229–257 Sunderland, MA: Sinauer Press [Google Scholar]
- 16.Roman J., Darling J. A. 2007. Paradox lost: genetic diversity and the success of aquatic invasions. Trends Ecol. Evol. 22, 454–464 10.1016/j.tree.2007.07.002 (doi:10.1016/j.tree.2007.07.002) [DOI] [PubMed] [Google Scholar]
- 17.Bernardi G., Golani D., Azzurro E. 2010. The genetics of Lessepsian bioinvasions. In Fish invasions of the Mediterranean Sea: change and renewal (eds Golani D., Appelbaum-Golani B.), pp. 71–84 Sofia, Bulgaria: Pensoft Publishers [Google Scholar]
- 18.Hassan M., Harmelin-Vivien M., Bonhomme F. 2003. Lessepsian invasion without bottleneck: example of two rabbitfish species (Siganus rivulatus and Siganus luridus). J. Exp. Mar. Biol. Ecol. 291, 219–232 10.1016/S0022-0981(03)00139-4 (doi:10.1016/S0022-0981(03)00139-4) [DOI] [Google Scholar]
- 19.Lockwood J. L., Casse P., Blackburn T. 2005. The role of propagule pressure in explaining species invasions. Trends Ecol. Evol. 20, 223–228 10.1016/j.tree.2005.02.004 (doi:10.1016/j.tree.2005.02.004) [DOI] [PubMed] [Google Scholar]
- 20.Kolbe J. J., Glor R. E., Schettino L. R., Lara A. C., Larson A., Losos J. B. 2004. Genetic variation increases during biological invasion by a Cuban lizard. Nature 431, 480–483 10.1038/nature02807 (doi:10.1038/nature02807) [DOI] [PubMed] [Google Scholar]
- 21.Genton B. J., Shykoff A., Giraud T. 2005. High genetic diversity in French invasive populations of common ragweed, Ambrosia artemisiifolia, as a result of multiple sources of introduction. Mol. Ecol. 14, 4275–4285 10.1111/j.1365-294X.2005.02750.x (doi:10.1111/j.1365-294X.2005.02750.x) [DOI] [PubMed] [Google Scholar]
- 22.Rosenthal D. M., Ramakrishnan A. P., Cruzan M. B. 2008. Evidence for multiple sources of invasion and intraspecific hybridization in Brachypodium sylvaticum (Hudson) Beauv. in North America. Mol. Ecol. 17, 4657–4669 10.1111/j.1365-294X.2008.03844.x (doi:10.1111/j.1365-294X.2008.03844.x) [DOI] [PubMed] [Google Scholar]
- 23.Randall J. E., Kanayama R. K. 1972. Hawaiian fish immigrants. Sea Front. 18, 144–153 [Google Scholar]
- 24.Oda D. K., Parrish J. D. 1982. Ecology of commercial snappers and groupers introduced to Hawaiian reefs. In Proc. 4th Int. Coral Reef Symp vol. 1, pp 59–67 Manila, Philippines: International Society for Reef Studies; University of the Philippines, Manila. 18–22 May 1981. [Google Scholar]
- 25.Randall J. E. 1987. Introductions of marine fishes to the Hawaiian Islands. Bull. Mar. Sci. 41, 490–502 [Google Scholar]
- 26.Meyer A. L., Dierking J. 2011. Elevated size and body condition and altered feeding ecology of the grouper Cephalopholis argus in non-native habitats. Mar. Ecol. Prog. Ser. 439, 203–212 10.3354/meps0933 (doi:10.3354/meps0933) [DOI] [Google Scholar]
- 27.Vignon M., Sasal P., Rigby M. C., Galzin R. 2009. Multiple parasite introduction and host management plan: case study of lutjanid fish in the Hawaiian Archipelago. Dis. Aquat. Org. 85, 133–145 10.3354/dao02071 (doi:10.3354/dao02071) [DOI] [PubMed] [Google Scholar]
- 28.Vignon M., Sasal P., Galzin R. 2009. Host introduction and parasites: a case study on the parasite community of the peacock grouper Cephalopholis argus (Serranidae) in the Hawaiian Islands. Parasitol. Res. 104, 775–782 10.1007/s00436-008-1254-3 (doi:10.1007/s00436-008-1254-3) [DOI] [PubMed] [Google Scholar]
- 29.Gaither M. R., Bowen B. W., Toonen R. J., Planes S., Messmer V., Earle J., Robertson D. R. 2010. Genetic consequences of introducing two allopatric lineages of Bluestriped Snapper (Lutjanus kasmira) to Hawaii. Mol. Ecol. 19, 1107–1121 10.1111/j.1365-294X.2010.04535.x (doi:10.1111/j.1365-294X.2010.04535.x) [DOI] [PubMed] [Google Scholar]
- 30.Randall J. E. 2007. Reef and shore fishes of the Hawaiian Islands. Honolulu, HI: Sea Grant College Program, University of Hawaii [Google Scholar]
- 31.Gaither M. R., Toonen R. J., Robertson D. R., Planes S., Bowen B. W. 2010. Genetic evaluation of marine biogeographic barriers: perspectives from two widespread Indo-Pacific snappers (Lutjanus kasmira and Lutjanus fulvus). J. Biogeog. 37, 133–147 10.1111/j.1365-2699.2009.02188.x (doi:10.1111/j.1365-2699.2009.02188.x) [DOI] [Google Scholar]
- 32.Gaither M. R., Bowen B. W., Bordenave T., Rocha L. A., Newman S. J., Gomez J. A., van Herwerden L., Craig M. T. 2011. Phylogeography of the reef fish Cephalopholis argus (Epinephelidae) indicates Pleistocene isolation across the Indo-Pacific Barrier with contemporary overlap in the Coral Triangle. BMC Evol. Biol. 11, 189. 10.1186/1471-2148-11-189 (doi:10.1186/1471-2148-11-189) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seutin G., White B. N., Boag P. T. 1991. Preservation of avian and blood tissue samples for DNA analyses. Can. J. Zool. 69, 82–92 10.1139/z91-013 (doi:10.1139/z91-013) [DOI] [Google Scholar]
- 34.Gaither M. R., Szabó Z., Crepeau M. W., Bird C. E., Toonen R. J. 2011. Preservation of corals in salt-saturated DMSO buffer is superior to ethanol for PCR experiments. Coral Reefs 30, 329–333 10.1007/s00338-010-0687-1 (doi:10.1007/s00338-010-0687-1) [DOI] [Google Scholar]
- 35.Nei M. 1987. Molecular evolutionary genetics. New York, NY: Columbia University Press [Google Scholar]
- 36.Excoffier L., Lischer H. E. L. 2010. Arlequin suite v. 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Res. 564–567 10.1111/j.1755-0998.2010.02847.x (doi:10.1111/j.1755-0998.2010.02847.x) [DOI] [PubMed] [Google Scholar]
- 37.Bandelt H. J., Forster P., Röhl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48 10.1093/oxfordjournals.molbev.a026036 (doi:10.1093/oxfordjournals.molbev.a026036) [DOI] [PubMed] [Google Scholar]
- 38.Leberg P. L. 2002. Estimating allelic richness: Effects of sample size and bottlenecks. Mol. Ecol. 11, 2445–2449 10.1046/j.1365-294X.2002.01612.x (doi:10.1046/j.1365-294X.2002.01612.x) [DOI] [PubMed] [Google Scholar]
- 39.Bowen B. W., Bass A. L., Rocha L. A., Grant W. S., Robertson D. R. 2001. Phylogeography of the trumpetfish (Aulostomus spp.): ring species complex on a global scale. Evolution 55, 1029–1039 10.1554/0014-3820(2001)055[1029:POTTAR]2.0.CO;2 (doi:10.1554/0014-3820(2001)055[1029:POTTAR]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 40.Lessios H. A. 2008. The great American schism: divergence of marine organisms after the rise of the Central American Isthmus. Annu. Rev. Ecol. Evol. Syst. 39, 63–91 10.1146/annurev.ecolsys.38.091206.095815 (doi:10.1146/annurev.ecolsys.38.091206.095815) [DOI] [Google Scholar]
- 41.Reece J. S., Bowen B. W., Joshi K., Goz V., Larson A. F. 2010. Phylogeography of two moray eels indicates high dispersal throughout the Indo-Pacific. J. Hered. 101, 391–402 10.1093/jhered/esq036 (doi:10.1093/jhered/esq036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Randall J. E., Earle J. L., Pyle R. L., Parrish J. D., Hayes T. 1993. Annotated checklist of the fishes of Midway Atoll, Northwestern Hawaiian Islands. Pac. Sci. 47, 356–400 [Google Scholar]
- 43.Martel S. J. D., Korman J., Darcy M., Christensen L. B., Zeller D. 2006. Status and trends of the Hawaiian bottomfish stocks: 1948–2004. Vancouver, Canada: University of British Columbia Fisheries Centre [Google Scholar]
- 44.Excoffier L., Foll M., Petit R. J. 2009. Genetic consequences of range expansions. Annu. Rev. Ecol. Evol. Syst. 40, 481–501 10.1146/annurev.ecolsys.39.110707.173414 (doi:10.1146/annurev.ecolsys.39.110707.173414) [DOI] [Google Scholar]
- 45.Toonen R. J., et al. 2011. Defining boundaries for ecosystem-based management: a multispecies case study of marine connectivity across the Hawaiian Archipelago. J. Mar. Biol. 2011, 460173 10.1155/2011/460173 (doi:10.1155/2011/460173) [DOI] [PMC free article] [PubMed] [Google Scholar]