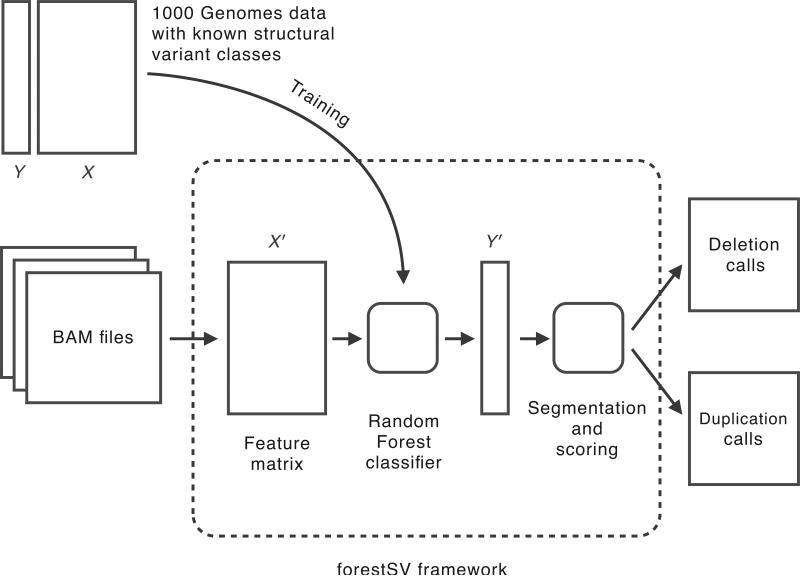
Figure 1. The forestSV framework.
The core of the framework is a Random Forest classifier trained to recognize structural variants based on calls and data (Y and X, respectively) from the 1000 Genomes Project. To make calls on new data, BAM files are given as input. The data from the BAM files are used to construct a new feature matrix X'. The classifier provides a mapping from X' to predicted structural variant classes, Y'. The predictions are segmented into structural variant calls and scored according to the confidence in their class assignment. The output of the framework is a pair of lists of predicted deletion and duplication events, with their associated confidence scores.