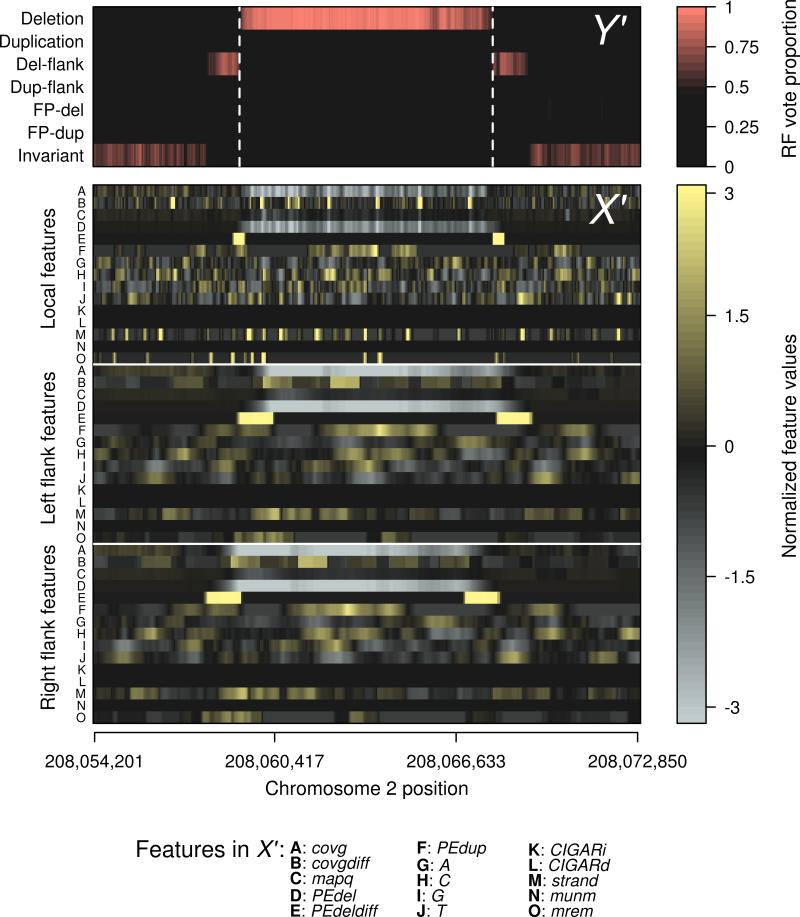
Figure 2. Mapping of features to structural variant calls by Random Forests.
Sequence features (A-O in matrix X') from a representative deletion locus on chromosome 2 of the autistic individual 03C14438, and their corresponding classification (Y') by the classifier as a deletion event. The estimated boundaries of the deletion event are shown by the dashed lines. Because the classifier has been trained to recognize regions that flank known structural variants, the presence of these landmarks, independent of the deletion call itself, provides additional support for the presence of an event. Features are labeled according to their designation given in the forestSV package, and are also described in methods. Detailed descriptions of the features are provided in Supplementary Table 2. Features have been standardized to mean 0 and variance 1.