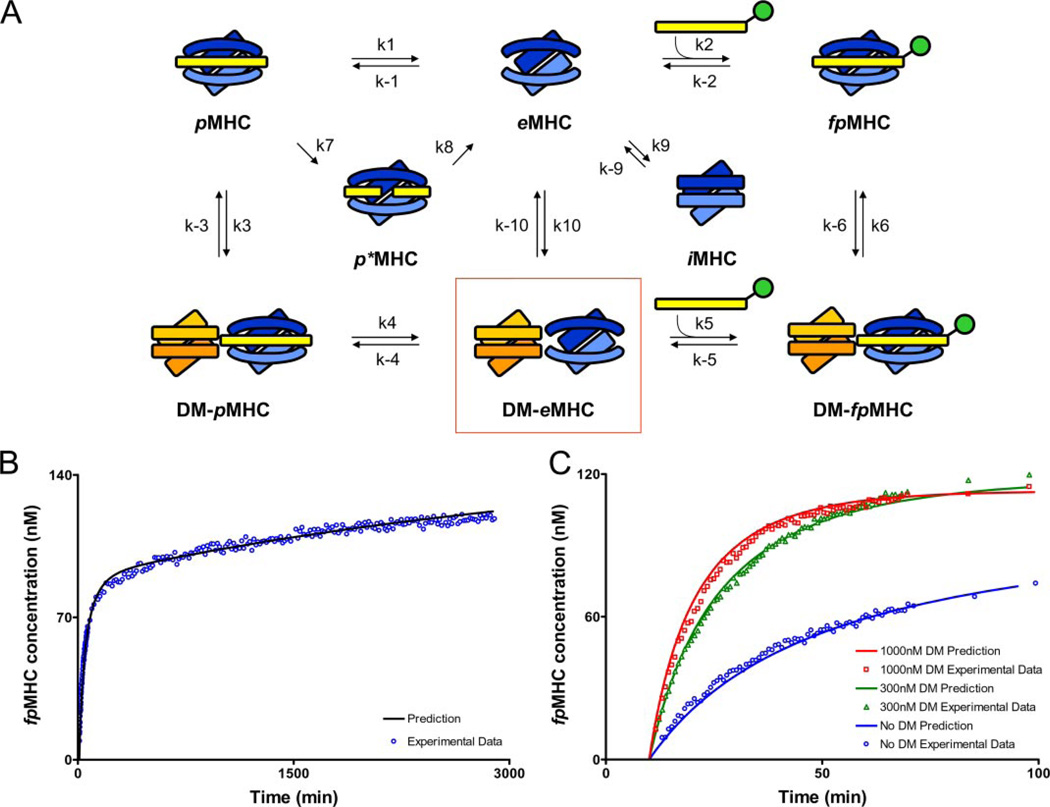
FIGURE 7. Model showing the rate constants used in mathematical models of peptide loading in the presence and absence of DM.
A, peptide-loaded MHC (pMHC), empty MHC (eMHC), and fluorescent peptide-loaded MHC (fpMHC) are shown in the absence of DM (top row) and interacting with DM (bottom row). Peptide photocleavage creates a short-lived MHC molecule with bound cleavage fragments (p*MHC), whereas incubation in the absence of peptide creates an inactive form (iMHC). eMHC remains in a peptide-receptive form only for short periods of time, but eMHC bound in the DM-eMHC complex (boxed) remains active for extended periods of time. B, the hypothesis represented by Equation 2 fits the experimental data without DM very well. The initial concentration of pMHC was 150 nm, and the amount of MBP-488 added after 10 min of UV was 200 nm. C, the model represented by Equation 3 fits the experimental data well in the presence of variable amounts of DM. The initial concentration of pMHC was 150 nm, and the amount of MBP-488 added after 10 min of UV was 200 nm. DM addition occurred at the same time as MBP-488 addition. A summary of the concordance between experimental data and the kinetic model for conditions not shown here is provided in the supplemental information. Because of uncertainties associated with the length of time required to add MBP-488 and to take the first reading, it was assumed that 30 s elapsed during this process for all DM experiments. Because the no DM data were read last, it was assumed that 2 min elapsed before the first reading. The values of the delay times were chosen to improve the quality of the fit. We also carried out the analyses assuming that the elapsed times were negligible. None of the qualitative conclusions changed upon making this assumption; only the quantitative comparisons with experimental data were less precise at short times.