Abstract
Alkaptonuria (AKU) is a rare genetic disease associated with the accumulation of homogentisic acid (HGA) and its oxidized/polymerized products which leads to the deposition of melanin-like pigments (ochronosis) in connective tissues. Although numerous case reports have described ochronosis in joints, little is known on the molecular mechanisms leading to such a phenomenon. For this reason, we characterized biochemically chondrocytes isolated from the ochronotic cartilage of AKU patients. Based on the macroscopic appearance of the ochronotic cartilage, two sub-populations were identified: cells coming from the black portion of the cartilage were referred to as “black” AKU chondrocytes, while those coming from the white portion were referred to as “white” AKU chondrocytes. Notably, both AKU chondrocytic types were characterized by increased apoptosis, NO release, and levels of pro-inflammatory cytokines. Transmission electron microscopy also revealed that intracellular ochronotic pigment deposition was common to both “white” and “black” AKU cells. We then undertook a proteomic and redox-proteomic analysis of AKU chondrocytes which revealed profound alterations in the levels of proteins involved in cell defence, protein folding, and cell organization. An increased post-translational oxidation of proteins, which also involved high molecular weight protein aggregates, was found to be particularly relevant in “black” AKU chondrocytes. J. Cell. Physiol. 227: 3333–3343, 2012. © 2011 Wiley Periodicals, Inc.
Alkaptonuria (AKU; MIM no. 203500) is an autosomal recessive inherited rare disease resulting from a deficiency of the enzyme homogentisate 1,2-dioxygenase (HGO; EC 1.13.11.) that normally splits the aromatic ring of homogentisic acid (HGA; 2,5-dihydroxyphenylacetic acid), an intermediary product of the catabolism of tyrosine and phenylalanine (La Du et al., 1958; Fernandez-Canon et al., 1996; Phornphutkul et al., 2002). As a consequence, HGA is not further metabolized and is excreted with urine, to which it imparts a black discoloration upon standing due to its oxidation to benzoquinone acetic acid (BQA) (Zannoni et al., 1962). The same oxidation occurs within the body, where HGA is accumulated, allowing the formation of oxidized/polymerized melanin-like products imparting to connective tissues (especially skin, cardiovascular system, and joints) a characteristic pathologic pigmentation known as “ochronosis” (Zannoni et al., 1962; O'Brien et al., 1963; Phornphutkul et al., 2002; Helliwell et al., 2008). Ochronosis leads to rapid degeneration and inflammation of joints where a concomitant musculoskeletal involvement might induce a severe and sometimes crippling form of arthropathy (Selvi et al., 2000; Helliwell et al., 2008).
Yet well described from the clinical point of view, alkaptonuric ochronosis and its molecular mechanisms have not been explored to any significant degree due mainly to the rarity of the disease [AKU has a 1:250,000–1,000,000 incidence (Tinti et al., 2011a)] as well as to the lack of suitable models to study it. We recently introduced novel human ochronotic cell and serum models in which we demonstrated how HGA induces protein oxidation and aggregation (Braconi et al., 2010a, b; Tinti et al., 2010, 2011b). In the present paper, we provided the first proteomic characterization of AKU chondrocytes, obtained from the ochronotic cartilage of AKU patients. Our findings indicated that AKU cells experience inflammatory stimuli and are characterized by relevant alterations in the expression of proteins involved in cell defence, protein folding and cell organization, sharing similarities with osteoarthritis (OA). Additionally, AKU cells experience significant protein oxidation and aggregation which might help, in turn, the production of ochronotic pigments.
Materials and Methods
Reagents
Unless otherwise indicated, all high quality reagents and antibodies were from Sigma–Aldrich (St. Louis, MO). All water used was Milli-Q (Millipore, Bedford, MA).
Isolation and culture of human chondrocytes from ochronotic AKU patients
AKU chondrocytes were obtained, after informed consent in accordance with the Declaration of Helsinki, from hip cartilage fragments of three alkaptonuric patients suffering from ochronotic arthropathy and undergone surgery for hip replacement. The study received approval from the Local Ethics Committee. The characteristics of patients enrolled for the study are schematically reported below:
Patient #1: female, age 62, 4/4 backbone impairment, 4/4 articular joints impairment, two orthopaedic surgical interventions, urinary HGA level (24 h): 300 mg/dl.
Patient #2: male, age 60, 4/4 backbone impairment, 4/4 articular joints impairment, five orthopaedic surgical interventions, urinary HGA level (24 h): 371 mg/dl.
Patient #3: female, age 69, 4/4 backbone impairment, 4/4 articular joints impairment, two orthopaedic surgical interventions, urinary HGA level (24 h): 475 mg/dl.
At visual inspection, donors' cartilages presented both a white and dark coloration and we obtained, from separate digestion of these different fragments, two cell populations that we named “white” and “black” chondrocytes. Immediately after surgery, cartilage was cut aseptically and minced in small pieces. Fragments were washed in Dulbecco's modified Eagle's medium (DMEM) containing 2% penicillin/streptomycin solution and 0.2% amphoterycin B. Chondrocytes were isolated by sequential enzymatic digestion: 30 min with 0.1% hyaluronidase, 1 h with 0.5% pronase, and 1 h with 0.2% collagenase at 37 °C in wash solution (DMEM + penicillin/streptomycin solution + amphoterycin B). The cells suspension was then filtered twice using 70 µm nylon meshes, washed and centrifuged for 10 min at 700×g. Cell pellets were then re-suspended in DMEM containing 10% fetal calf serum (FCS) and 0.33 mM HGA and expanded in monolayer culture at 37°C in 95% relative humidity and in a 5% CO2 atmosphere. HGA was added to mimic in vitro the AKU pathological condition; the concentration used, 0.33 mM, is in the range of circulating HGA in AKU patients' serum (Angeles et al., 1989) and it has been successfully used in several AKU models that we have set up and characterized previously (Braconi et al., 2010a, b; Tinti et al., 2010, 2011a, b). In parallel, control (non-AKU) chondrocytes from OA samples were obtained and cultured under the same conditions but avoiding HGA addition.
Cell proliferation
The DNA content of cell pellets from each culture was used as a parameter of cell proliferation. DNA was extracted with a high pure PCR template preparation kit according to manufacturer's instructions (Roche Diagnostics, Mannheim, Germany) and quantified with Qubit™ (Invitrogen, Grand Island, NY).
Apoptosis
Apoptosis was analyzed by flow cytometry evaluating Annexin V binding (DAKO, Milano, Italy). Cells were stained with fluorescein isothiocyanate (FITC)-Annexin V and propidium iodide.
NO release
Nitrite levels were measured in chondrocyte culture supernatants with Griess reagent (1% sulfanilamide, 0.1% napthylenediamine dihydrochloride, and 2.5% phosphoric acid) (Rediske et al., 1994). Equals volumes (100 µl) of supernatant and Griess reagent were incubated in microtitre plates at room temperature for 5 min. Nitrite concentrations were determined spectrophotometrically at 550 nm using a standard curve generated with serial dilutions of sodium nitrite.
Transmission electron microscopy (TEM)
Once isolated from ochronotic specimens, AKU chondrocytes were fixed for 2 h at 4°C in cold Karnovsky fixative, rinsed overnight in 0.1 M pH 7.2 cacodylate buffer, post-fixed for 1 h at 4°C in 1% buffered OsO4, dehydrated in a graded series of ethanol, and embedded in Epon-Araldite. Ultrathin sections cut with an LKB III ultramicrotome were mounted on copper grids, stained with uranyl acetate and lead citrate and then photographed with a Philips CM10 electron microscope. At least 100 cells from each group were observed.
Cytokine assay
The release of IL-1β, IL-6 IL-8, IL-10, and TNFα from AKU chondrocytes was evaluated in cell culture supernatant by means of multiplex assay for cytokine quantification (Bioplex, Bio-Rad, Hercules, CA). Cytokine concentrations were calculated using a standard curve established from serial dilutions of each cytokine standard as described in the manufacturer's protocol and expressed as picograms per milliliter of culture medium.
Two-dimensional electrophoresis (2D-PAGE) and Western blot
Cells were washed twice with sterile PBS and resuspended in 50 µl of a buffer containing 65 mM DTE, 65 mM CHAPS, 9 M urea, and 35 mM Tris-base. Cell disruption was achieved by sonicating briefly in an ice bath. Protein content in cell lysates was assessed according to Bradford (Bradford, 1976).
Cell lysates were first mixed with a buffer containing 8 M urea, 35 mM CHAPS, 10 mM DTE, and a trace of bromophenol blue. One hundred micrograms [2D gels to be transferred onto nitrocellulose (NC) membranes] or 50 µg (2D gels to be silver stained) of proteins were adsorbed onto Immobiline Dry Strips (IPG 18 cm, nonlinear 3–10 pH range, Bio-Rad) for 10 h. Isoelectric focusing (IEF) was carried out with a Protean IEF cell (Bio-Rad). The voltage was linearly increased from 300 to 3,500 V during the first 3 h and then stabilized at 5,000 V for 22 h (total 110 kV × h).
Prior to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE), IPG were equilibrated in 6 M urea, 30% (v/v) glycerol, 2% (w/v) SDS, 0.05 M Tris–HCl pH 6.8 containing first 2% (w/v) DTE, and later 2.5% (w/v) iodoacetamide (IAA). SDS–PAGE was carried out applying 40 V per gel until the dye front reached the bottom of gels.
Silver ammoniacal staining was carried out according to Switzer (Switzer et al., 1979) whereas gels to be transferred onto NC were washed and equilibrated in a transfer buffer [50 mM Tris, 40 mM glycine, 1.3 mM SDS, 20%, v/v methanol] and protein transfer was carried out using a semidry Novablot transblot cell (Bio-Rad) applying 0.7 mA/cm2 for 75 min. Protein transfer was checked by staining with 0.2% Ponceau S in 3% (v/v) TFA for 3 min and de-staining with water.
For the Western blot analysis of carbonylated proteins, after the IEF IPG strips were briefly rinsed with water and incubated at room temperature for 20 min with 10 mM 2,4-dinitrophenylhydrazine (DNPH) in 5% (v/v) TFA to allow derivatization of protein carbonyls (Reinheckel et al., 2000). Strips were washed twice with a solution containing 8 M urea, 20% (v/v) glycerol, 9 M SDS, and 150 mM Tris–HCl pH 6.8. For the immunorevelation of protein carbonyls, NC sheets were incubated with rabbit anti-dinitrophenyl antibodies 1:10,000 (over-night at 4°C), followed by peroxidase-conjugated anti-rabbit antibodies 1:7,000 (2 h at room temperature) and revelation was achieved through chemiluminescence (Immu-Star HRP, Bio-Rad).
Protein spots of interest were identified by MALDI-TOF mass spectrometry (MS) or by gel matching with proteomic reference maps already produced, calibrated, and characterized in our laboratories (Spreafico et al., 2006, 2009).
Image analysis
Images of gels and films were acquired (Image Scanner, Amersham Biosciences, Uppsala, Sweden) and analyzed with Image Master™ Platinum (Amersham Biosciences). For comparative proteomic analysis, spot% relative abundance was adopted; for multiple spots identified as different molecular species of a same protein, the average% relative abundance was calculated. For the quantitative analysis of immunorevealed protein carbonyls, the intensity of bands, which is automatically normalized by Image Master™ Platinum against the surrounding background, was chosen as the reference parameter.
Statistical analysis
All of the experiments were carried out in triplicate; data are presented as average values with standard deviation. Student's t-test and multiple-measurement ANOVA analysis followed by the Bonferroni-type multiple comparison were used when necessary. Differences with at least a P-value ≤0.05 were considered significant. For comparative proteomic and redox proteomic analyses, only representative gels and films are shown.
Results
In this work, we characterized two distinct types of primary chondrocytes (namely “white” and “black” AKU chondrocytes) obtained from AKU patients suffering from ochronotic arthropathy who underwent surgery. TEM analysis revealed that both “white” and “black” AKU chondrocytes contained intracellular ochronotic pigments (Fig. 1) and showed a similar morphology. The finding of pigments in “white” chondrocytes is particularly noteworthy, because it demonstrated that deposition of ochronotic pigments occurred even in areas where no macroscopic evidence of pigmentation was observed (Kutty et al., 1973). Therefore, intracellular pigment deposition possibly precedes deposition in the extracellular matrix. Another noteworthy feature was the presence of ochronotic deposits of varying shapes and sizes also within some vacuoles (Fig. 1).
Fig. 1.
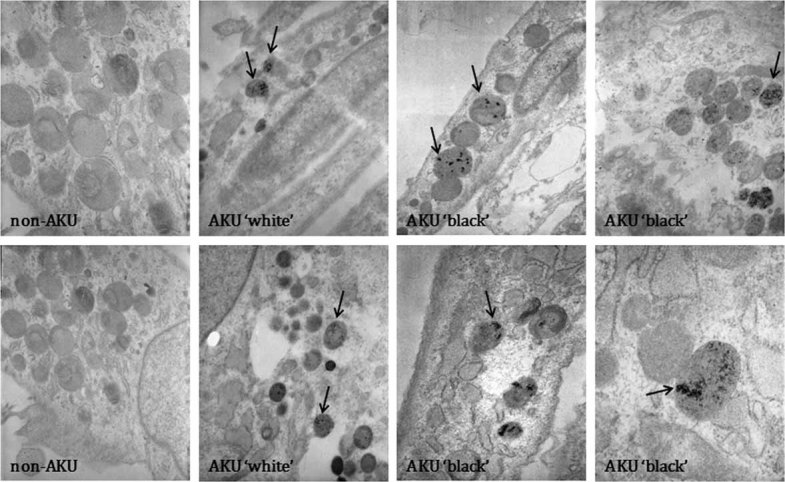
Deposition of ochronotic pigments, detected by TEM, in “white” and “black” AKU chondrocytes and their controls (non-AKU). Arrows indicate the presence of ochronotic pigments. Only representative images are shown.
Both types of AKU cells showed a proliferation rate higher than the control (+30.8- and +8.9-fold-change for “white” and “black” chondrocytes, respectively) and “white” AKU chondrocytes had a proliferation rate higher than “black” ones (with a +6.1-fold change) (Fig. 2A). Since exposure to HGA may induce cytotoxicity (Kirkpatrick et al., 1984; Angeles et al., 1989), we investigated also cell apoptosis and found that “white” and “black” chondrocytic cultures were both characterized by a higher% of apoptotic cells when compared to the control (+1.5- and +2.0-fold-change, respectively) (Fig. 2B). Interestingly, the release of NO in AKU chondrocytes was significantly higher than in the control. Particularly, +33- and +126-fold-change values were recorded for NO levels in “white” and “black” chondrocytes, respectively (Fig. 2C).
Fig. 2.
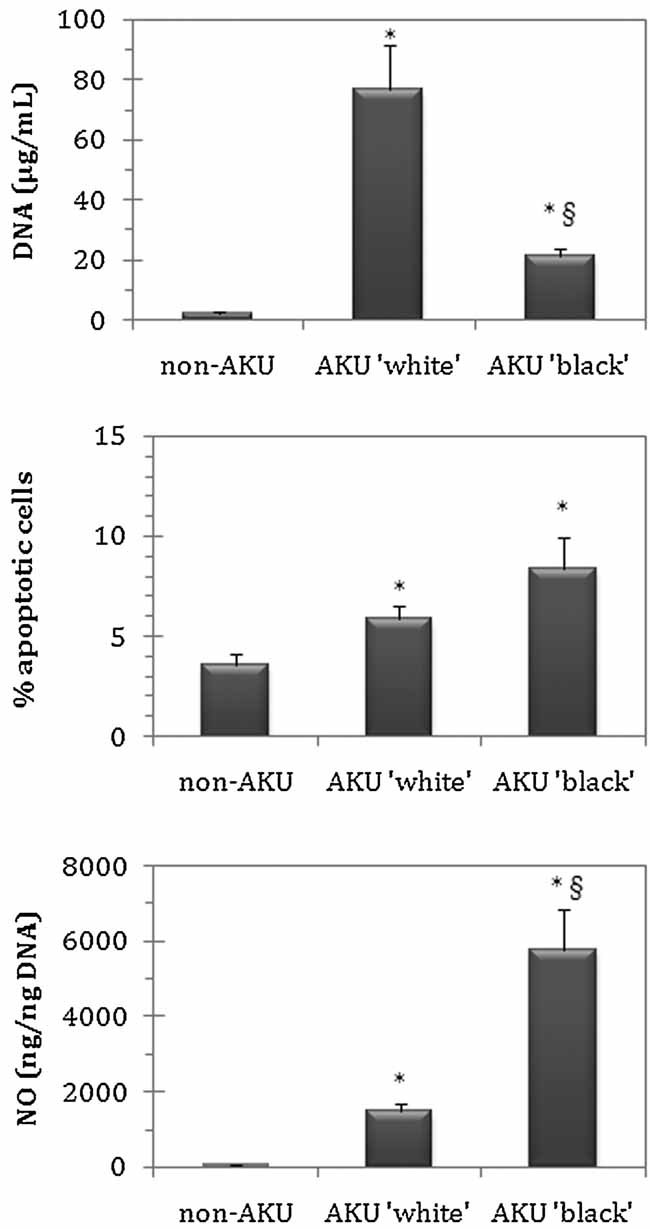
Cell proliferation (A), apoptosis (B), and NO release (C) in “white” and “black” AKU chondrocytes and their control (non-AKU). Cell proliferation was assayed by measuring the DNA content of cell pellets; apoptosis was assayed by Annexin V-FITC/propidium iodide staining and flow cytometry; NO release in culture supernatants was assayed by Griess reagent, as detailed under Materials and Methods Section. Experiments were performed in triplicate; data are presented as average values with standard deviation. Statistical significance compared to non-AKU control (*P ≤ 0.05) and between “white” and “black” AKU chondrocytes (§P ≤ 0.05) is indicated.
Cytokine expression
To evaluate the inflammatory status of AKU ochronotic chondrocytes, we measured the levels of IL-1β, IL-6, IL-8, IL-10, and TNFα in cell culture supernatants. With the only exception of IL-1β in “black” AKU chondrocytes, which did not show any difference with respect to the control, for each assayed cytokine we found significantly higher levels in AKU cells, as shown in Figure 3. Interestingly, levels of both IL-1β and TNFα were significantly higher in “white” AKU chondrocytes, whereas “black” chondrocytes showed only a significant increase in TNFα levels. Upon injuries or diseases, cytokines such as IL-1β and TNFα can be found as major mediators of inflammation within articular cartilages undergoing repair/regeneration (Pelletier et al., 2001; Kobayashi et al., 2005; Wehling et al., 2009). Both cytokines induce an imbalance towards the catabolic pathway (Fernandes et al., 2002) through up-regulation of metalloproteinases (MMPs), aggrecanases, inducible NO synthase, and cyclooxygenase 2 (Umlauf et al., 2010). Their pro-inflammatory action is also mediated by an enhanced production of IL-6 and IL-8 (Umlauf et al., 2010) together with induction of apoptosis (Fischer et al., 2000; Aizawa et al., 2001; Aigner and Kim, 2002). Though players in a very complex scenario, IL-1β and TNFα are associated with cartilage degradation and synovial inflammation observed in OA (Fernandes et al., 2002).
Fig. 3.
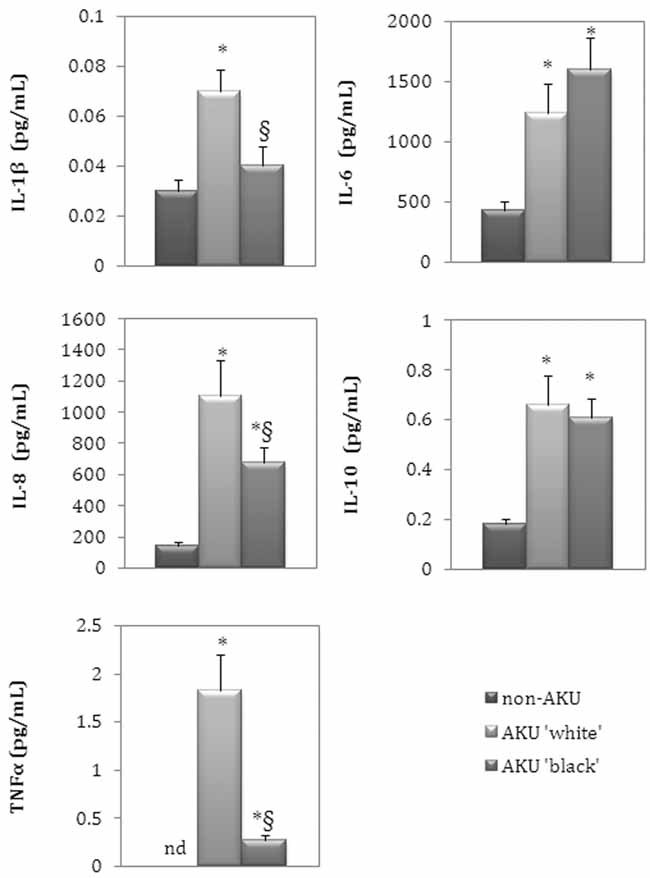
Release of cytokines in “white” and “black” AKU chondrocytes and their control (non-AKU) by means of multiplex assay, as detailed under Materials and Methods Section. Experiments were performed in triplicate; data are presented as average values with standard deviation. Statistical significance compared to non-AKU control (*P ≤ 0.05) and between “white” and “black” AKU chondrocytes (§P ≤ 0.05) is indicated. nd: non-detectable.
IL-6 levels were significantly increased in AKU “white” and “black” chondrocytes with respect to the control (+2.9- and +3.7-fold-change, respectively), similarly to what we observed for IL-8 levels (+7.7-fold-change in “white” and +4.7-fold-change in “black” AKU chondrocytes). IL-6 is a co-factor of the catabolic effects of IL-1β (Flannery et al., 2000) enhancing the production of MMP-13 and, consequently, the IL-1β-induced degradation of proteoglycan in cartilage (Honorati et al., 2002). IL-8 belongs to the family of chemokines. Recently, a key role for the chondrocytic chemokine/chemokine receptor system in OA has been suggested (Facchini et al., 2005). Notably, IL-8 was used as a biomarker for rheumatoid arthritis on the basis of its elevated levels found in synovial fluid of affected patients (Kokebie et al., 2011).
IL-10 levels were 3.7 and 3.4 times higher in AKU “white” and “black” chondrocytes than in the control, respectively. In OA, cartilage and synovium show increased levels of IL-10 (Isomaki and Punnonen, 1997; Jorgensen et al., 1998; Moo et al., 2001) that are thought to inhibit the synthesis of other pro-inflammatory cytokines (Katsikis et al., 1994; Lechman et al., 1999). Nevertheless, the exact roles of IL-10 in inflammatory diseases, and especially in chondrocyte homeostasis, need to be fully realized. In particular, the study of the interplay between IL-10 and TNFα revealed that, if on the one hand IL-10 can strongly inhibit the TNFα-induced down regulation of aggrecan expression, on the other it can only have minor effects in contrasting the TNFα-induced collagen type II suppression (Muller et al., 2008).
Comparative proteomics
Silver staining allowed to reveal nearly 3,000 spots in “white” and “black” AKU chondrocyte proteomic maps (Fig. 4A, B). MALDI-ToF MS and gel matching with reference maps (Spreafico et al., 2006, 2009) allowed the identification of 78 gene products. Once set a statistically significant threshold ≥2.0 for fold-change values in protein relative abundance ratio, the quantitative analysis against the control proteomic map revealed that 34 and 41 proteins were differently expressed in “white” and “black” AKU chondrocytes, respectively. The description of differently expressed proteins and fold-change values are schematically reported in Table I (see also Supplementary Material). The functional classification of differently expressed proteins is reported in Figure 4C, D. The protein repertoires of “white” and “black” AKU chondrocytes were quite similar and the most significant alterations with respect to the control were found for proteins involved in:
Fig. 4.
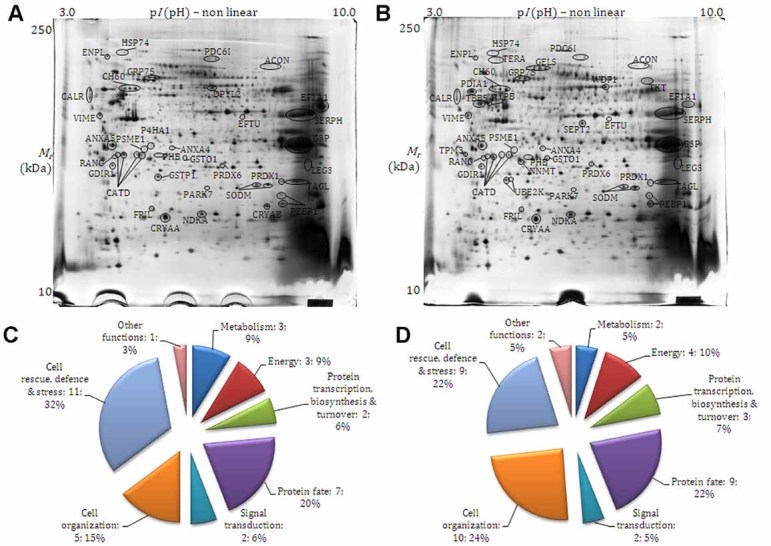
Proteomic analysis. Silver stained 2D maps of “white” (A) and “black” (B) AKU chondrocytes. The comparative analysis carried out against a non-AKU chondrocytes control map (not shown) allowed the identification of differently expressed proteins (fold change at least 2 in protein% relative abundance) that are indicated with their abbreviated name. Functional classification of differently expressed proteins is indicated in (C) and (D) for “white” and “black” AKU chondrocytes, respectively. Representative images from a triplicate set are shown.
Table I.
Comparative proteomics of AKU chondrocytes
| Spot | AN | Gene | Protein | Biological processesa | loc.b | CHW/CTRc | CHB/CTRc | CHB/CHW | IDd |
|---|---|---|---|---|---|---|---|---|---|
| Metabolism | |||||||||
| Nucleotide, nucleoside, and nucleic acids | |||||||||
| DPYL2 | Q16555 | DPYSL2 | Dihydropyrimidinase-related protein 2 | Signal transduction. Necessary for remodeling of the cytoskeleton | C, Cs | −2.8 | GM | ||
| NDKA | P15531 | NME1 | Nucleoside diphosphate kinase A | Nucleotide metabolism, cell differentiation, and regulation of apoptosis | C, N | +2.1 | +2.3 | GM | |
| PHB | P35232 | PHB | Prohibitin | Inhibition of DNA synthesis, regulation of proliferation | Mt | −11.9 | −3.2 | +3.7 | GM |
| Energy | |||||||||
| Carbohydrate metabolism | |||||||||
| ACON | Q99798 | ACO2 | Aconitate hydratase, mitochondrial | Tricarboxylic acid cycle; isocitrate from oxaloacetate: step 2/2 | Mt, N | +3.7 | +2.8 | GM | |
| G3P | P04406 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | Glycolysis (pyruvate from d-glyceraldehyde 3-phosphate: step 1/5). Independent of its glycolytic activity, it is also involved in membrane trafficking in the early secretory pathway and in oxido- reductase reactions | C, M | −5.2 | −3.9 | MS | |
| Respiration and fermentation | |||||||||
| ATPB | P06576 | ATP5B | ATP synthase subunit beta, mitochondrial | ATP synthesis, regulation of intracellular pH | Mt | −3.8 | −2.5 | GM | |
| LDHB | P07195 | LDHB | l-lactate dehydrogenase B chain | Fermentation; pyruvate fermentation to lactate; (S)-lactate from pyruvate: step 1/1 | C | −2.5 | — | — | GM |
| Pentose-phosphate pathway | |||||||||
| TKT | P29401 | TKT | Transketolase | Sedoheptulose 7-phosphate + d-glyceraldehyde 3-phosphate = d-ribose 5-phosphate + d-xylulose 5-phosphate; response to oxidative stress | C | +2.2 | +2.0 | GM | |
| Transcription, synthesis, and turnover of proteins | |||||||||
| CATD | P07339 | CTSD | Cathepsin D | Acid protease active in intracellular protein breakdown | Lys, Mel, S | −4.7 | −3.8 | MS | |
| EF1A1 | P68104 | EEF1A1 | Elongation factor 1-alpha 1 | Protein biosynthesis | C | −9.6 | −10.1 | GM | |
| EFTU | P49411 | TUFM | Elongation factor Tu, mitochondrial | Protein biosynthesis | Mt | −6.4 | −7.5 | GM | |
| Protein fate (folding, maturation, and transport) | |||||||||
| CALR | P27797 | CALR | Calreticulin | Molecular calcium-binding chaperone promoting folding, oligomeric assembly and quality control in the ER | C, ER, EM, S | −2.3 | −2.9 | GM | |
| GRP75 | P38646 | HSPA9 | 75 kDa glucose-regulated protein | Control of cell proliferation and cellular aging, may also act as a chaperone; has anti-apoptotic functions | Mt | −2.0 | −2.1 | GM | |
| P4HA1 | P13674 | P4HA1 | Prolyl 4-hydroxylase subunit alpha-1 | Catalyzes the post-translational formation of 4-hydroxyproline in -Xaa-Pro-Gly- sequences in collagens and other proteins; it is involved in redox reactions | −2.2 | GM | |||
| PARK7 | Q99497 | PARK7 | Protein DJ-1 | May function as a redox-sensitive chaperone and as a sensor for oxidative stress; prevents aggregation of alpha-synuclein | C, N, may be associated to Mt after oxidative stress | −3.3 | −3.4 | GM | |
| PDC6I | Q8WUM4 | PDCD6IP | Programmed cell death 6-interacting protein | Protein transport; may play a role in the regulation of both apoptosis and cell proliferation | C, Cs, Mel | +12.8 | +8.2 | −2.2 | GM |
| PDIA1 | P07237 | P4HB | Protein disulfide-isomerase | Catalyzes the formation, breakage, and rearrangement of disulfide bonds; at the cell surface, seems to act as a reductase that cleaves disulfide bonds of proteins attached to the cell. At high concentrations, functions as a chaperone that inhibits aggregation of misfolded proteins; at low concentrations, facilitates aggregation (anti-chaperone activity) | ER, Mel, M | −2.3 | GM | ||
| PEBP1 | P30086 | PEBP1 | Phosphatidylethanolamine-binding protein 1 | Phosphatidylethanolamine-binding protein; serine protease inhibitor | C | +16.4 | +27.2 | GM | |
| PSME1 | Q06323 | PSME1 | Proteasome activator complex subunit 1 | Implicated in immunoproteasome assembly and required for efficient antigen processing | C, Pr | −3.2 | −2.4 | GM | |
| TERA | P55072 | VCP | Transitional endoplasmic reticulum ATPase | ER-associated protein catabolic process, ER-unfolded protein response, protein ubiquitination, retrograde protein transport (ER to cytosol) | C, N | −2.9 | GM | ||
| UBE2K | P61086 | UBE2K | Ubiquitin-conjugating enzyme E2 K | Protein ubiquitination | C | −2.6 | GM | ||
| Signal transduction | |||||||||
| GDIR1 | P52565 | ARHGDIA | Rho GDP-dissociation inhibitor 1 | Regulates the GDP/GTP exchange reaction of the Rho proteins | C | −3.5 | −3.5 | GM | |
| RANG | P43487 | RANBP1 | Ran-specific GTPase-activating protein | Inhibits GTP exchange on Ran; may act in an intracellular signaling pathway which may control the progression through the cell cycle by regulating the transport of protein and nucleic acids across the nuclear membrane | C, N | −13.5 | −16.6 | GM | |
| Cellular organization | |||||||||
| Cytoskeleton and microtubules | |||||||||
| GELS | P06396 | GSN | Gelsolin | Binds to actin and to fibronectin preventing monomer exchange and promoting the assembly of monomers into filaments (nucleation) as well as sever filaments already formed. Defects in GSN are the cause of amyloidosis type 5 (AMYL5) [MIM:105120]; also known as familial amyloidosis Finnish type. AMYL5 is a hereditary generalized amyloidosis due to gelsolin amyloid deposition. It is typically characterized by cranial neuropathy and lattice corneal dystrophy. Most patients have modest involvement of internal organs, but severe systemic disease can develop in some individuals causing peripheral polyneuropathy, amyloid cardiomyopathy, and nephrotic syndrome leading to renal failure | C, Cs, S, amyloid | −2.0 | −2.2 | GM | |
| TAGL | Q01995 | TAGLN | Transgelin | Actin cross-linking/gelling protein. Involved in calcium interactions and contractile properties of the cell that may contribute to replicative senescence | C | +9.2 | +17.0 | MS | |
| TBB5 | P07437 | TUBB | Tubulin beta chain | Major constituent of microtubules | C | −2.9 | GM | ||
| Cell cycle | |||||||||
| SEPT2 | Q15019 | SEPT2 | Septin-2 | Cell division, mitosis | C | +2.0 | GM | ||
| Annexin family | |||||||||
| ANXA4 | P09525 | ANXA4 | Annexin A4 | Calcium/phospholipid-binding protein which promotes membrane fusion and is involved in exocytosis; anti-apoptosis | −4.9 | −2.4 | +2.0 | GM | |
| ANXA5 | P08758 | ANXA5 | Annexin A5 | Calcium-regulated membrane-binding protein, anti-apoptosis | C, N, M | −2.3 | −2.6 | GM | |
| Intermediate filaments family | |||||||||
| VIME | P08670 | VIM | Vimentin | Class-III intermediate filaments found in various non-epithelial cells | C | −5.2 | −4.1 | GM | |
| Other functions | |||||||||
| LEG3 | P17931 | LGALS3 | Galectin-3 | Cell differentiation | N, C, M | −3.3 | −11.3 | −3.4 | GM |
| TPM3 | P06753 | TPM3 | Tropomyosin alpha-3 chain | Stabilization of cytoskeleton and actin filaments, cell motion | C, Cs | −2.4 | GM | ||
| WDF1 | Q8IWB7 | WDFY1 | WD repeat and FYVE domain-containing protein 1 | Phosphatidylinositol binding | C, N | −2.6 | GM | ||
| Cell rescue, defence, and stress | |||||||||
| CH60 | P10809 | HSP60 | 60 kDa heat shock protein, mitochondrial | Implicated in mitochondrial protein import and macromolecular assembly, may facilitate the correct folding of imported proteins, prevent misfolding and promote the refolding and proper assembly of unfolded polypeptides generated under stress conditions in the mitochondrial matrix | Mt | −2.4 | −2.7 | GM | |
| CRYAA | P02489 | CRYAA | Alpha-crystallin A chain (Heat shock protein beta-4) | Anti-apoptosis, protein folding, unfolded protein binding, response to heat | C | +2.7 | +2.5 | GM | |
| CRYAB | P02511 | CRYAB | Alpha-crystallin B chain | Anti-apoptosis, protein folding, unfolded protein binding, response to heat | — | −2.2 | MS | ||
| ENPL | P14625 | HSP90B1 | Endoplasmin | Molecular chaperone that functions in the processing and transport of secreted proteins. Functions in endoplasmic reticulum associated degradation (ERAD). Has ATPase activity | C, ER, Mel | −3.6 | −4.8 | GM | |
| Plays a role in protein folding and transport; has anti-apoptotic functions; response to hypoxia | |||||||||
| GSTO1 | P78417 | GSTO1 | Glutathione S-transferase omega-1 | Exhibits glutathione-dependent thiol transferase and dehydroascorbate reductase activities | C | −5.2 | −2.2 | +2.3 | GM |
| RX + glutathione = HX + R-S-glutathione | |||||||||
| GSTP1 | P09211 | GSTP1 | Glutathione S-transferase P | Conjugation of reduced glutathione to a wide number of exogenous and endogenous hydrophobic electrophiles. It can have anti-apoptotic functions | C | −2.6 | MS | ||
| HSP74 | P34932 | HSPA4 | Heat shock 70 kDa protein 4 | Stress response; play a role in the unfolded protein response | C, N | −2.8 | −5.4 | GM | |
| PRDX1 | Q06830 | PRDX1 | Peroxiredoxin-1 | Cell redox-homeostasis, peroxide catabolic process, and cell proliferation | C, N, Mel | +7.6 | +8.2 | GM | |
| PRDX6 | P30041 | PRDX6 | Peroxiredoxin-6 | Cell redox homeostasis and protection against oxidative injury | C, N, L | P | P | GM | |
| SERPH | P50454 | SERPINH1 | Serpin H1 | Chaperone in the biosynthetic pathway of collage, it is induced by heat shock and plays a role in the response to unfolded protein | C | +2.7 | +3.1 | GM | |
| SODM | P04179 | SOD2 | Superoxide dismutase [Mn], mitochondrial | Destroys radicals which are normally produced within the cells and which are toxic to biological systems 2 superoxide + 2 H+ = O2 + H2O2 | Mt | −5.7 | −4.5 | GM | |
| Other functions | |||||||||
| FRIL | P02792 | FTL | Ferritin light chain | Iron storage and homeostasis, involved in redox reactions | R | +2.0 | +2.1 | GM | |
| NNMT | P40261 | NNMT | Nicotinamide N-methyltransferase | Catalyzes the N-methylation of nicotinamide and other pyridines to form pyridinium ions (this activity is important for biotransformation of many drugs and xenobiotic compounds) | C | −3.2 | −3.4 | GM | |
AN, accession number.
Proteins whose levels are altered in AKU “white” (CHW) and “black” (CHB) chondrocytes with respect to non-AKU control (CTR) chondrocytes.
Protein biological processes.
Protein sub-cellular localization: C (cytosol); Cs (cytoskeleton); CJ (cell junction); M (membrane); BM (basal membrane); EM (extracellular matrix); Mel (melanosome); Mt (mitochondrion); N (nucleus); Pe (peroxisome); Pr (proteasome); ER (endoplasmic reticulum); S (secreted); L (lysosome). Retrieved by UniProt knowledgebase (http://www.uniprot.org/).
Fold-change in protein% relative abundance (as average values in case of multiple spots); (+) over-expressed proteins, (−) under-expressed protein according to the ratio indicated. “A” and “P” indicate the absence or presence of proteins, respectively.
Identification method (GM: gel-matching; MS: MALDI-TOF mass spectrometry).
Protein fate: Twenty percent and 22% of proteins differently expressed in AKU “white” and “black” chondrocytes, respectively. We generally found an under expression of identified proteins involved in folding, maturation, and transport of proteins. It is noteworthy to mention the under-expression in “black” AKU chondrocytes of protein disulfide-isomerase (PDIA1), that is thought to contribute to the structural integrity of cartilage tissue, allowing a suitable response of load bearing joints to mechanical stress (Grimmer et al., 2006). We observed also an over-expression of programmed cell death 6-interacting protein (PDC6I) in both “white” and “black” AKU chondrocytes. Since PDC6I participates in the physiological sorting pathway for tyrosinase, a key enzyme in melanogenesis (Theos et al., 2005) an involvement of PDC6I in the production of ochronotic pigment in AKU at the cartilage level can be hypothesized.
Cell structure and organization: Fifteen percent and 24% of proteins differently expressed in AKU “white” and “black” chondrocytes, respectively. Changes in the organization and distribution of structural proteins characterize various pathologies, including OA (Fioravanti et al., 2003; Capin-Gutierrez et al., 2004; Holloway et al., 2004). Levels of several structural proteins were lower in AKU chondrocytes with respect to the control, such as vimentin (VIME), under-expressed in both “white” and “black” AKU chondrocytes, and gelsolin (GELS), under-expressed in “black” chondrocytes. VIME is the most sensitive sensor for mechanically induced stress, being necessary to maintain chondrocyte stiffness and allow a proper mechanotransduction (Li et al., 2010). GELS, regulating chondrocyte architecture and cell-matrix interactions, has been indicated as a prognostic and diagnostic marker of OA (De Ceuninck et al., 2005). Together, VIME and GELS contribute together to synovial fluid functions (Gobezie et al., 2007).
Cell rescue, defence, and stress response: Thirty-two percent and 22% of proteins differently expressed in AKU “white” and “black” chondrocytes, respectively. AKU chondrocytes showed altered levels of proteins fundamental for the protection from oxidative stress, like previously indicated by our group in a chondrocytic line used as a model of AKU (Braconi et al., 2010b). Though alpha-crystallin A chain (CRYAA), peroxiredoxin-1 (PRDX1), peroxiredoxin-6 (PRDX6), and serpin H1 (SERPH) were over-expressed both in “white” and “black” AKU chondrocytes compared to the control, lower protein levels were found in “white” and “black” AKU chondrocytes for most of the identified proteins. This is the case of the mitochondrial 60 kDa HSP (CH60), alpha-crystallin B chain (CRYAB), endoplasmin (ENPL), glutathione S-transferase omega-1 (GSTO1), glutathione S-transferase P (GSTP1), heat shock 70 kDa protein 4 (HSP74), and mitochondrial superoxide dismutase (SODM). The simultaneous deficiency of these proteins reinforce the hypothesis that AKU cells cannot adequately control reactive oxygen species (ROS)- and quinone-mediated toxicity.
Redox-proteomics
Differently from what observed with the comparative proteomic analysis, when we investigated protein oxidation through immunorevelation of carbonylated proteins we found that “white” and “black” AKU chondrocytes showed quite distinct patterns (Fig. 5). Gel-matching with the corresponding silver stained replica gel allowed the identification of almost all the spots immunorevealed under the conditions tested (Table II).
Fig. 5.
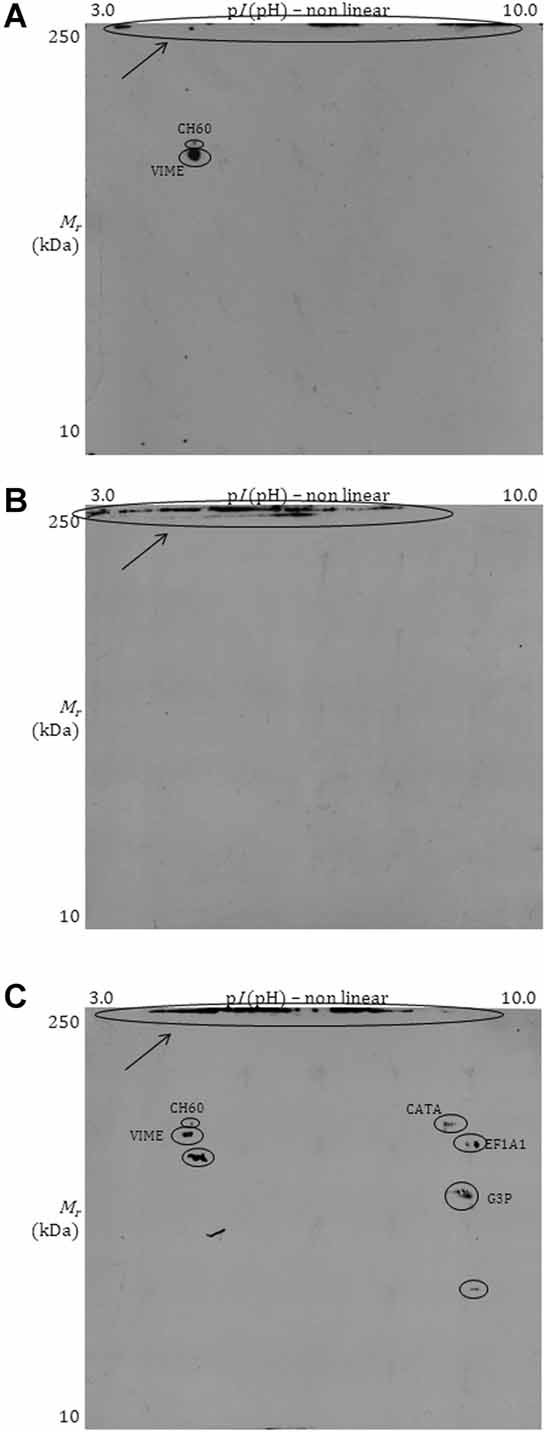
Redox proteomic analysis. Carbonylated proteins in non-AKU (A), “white” (B), and “black” (C) AKU chondrocytes are indicated with their abbreviated name. High molecular weight aggregates stacked on the top of the SDS–PAGE gel are highlighted with arrows. Representative images from a triplicate set are shown.
Table II.
Redox proteomics of AKU chondrocytes
| Spot | AN 1 | Gene | Protein | Biological processes 2 | CTR | CHW | CHB |
|---|---|---|---|---|---|---|---|
| Energy | |||||||
| Carbohydrate metabolism | |||||||
| G3P | P04406 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | Glycolysis, response to oxidative stress | + | + | |
| Transcription, synthesis, and turnover of proteins | |||||||
| EF1A1 | P68104 | EEF1A1 | Elongation factor 1-alpha 1 | Protein biosynthesis | + | ||
| Cellular organization | |||||||
| Intermediate filaments family | |||||||
| VIME | P08670 | VIM | Vimentin | Structural constituent of cytoskeleton, cell motion | + | + | |
| Cell rescue, defence, and stress | |||||||
| CATA | P04040 | CAT | Catalase | Response to oxidative stress | + | ||
| CH60 | P10809 | HSP60 | 60 kDa heat shock protein, mitochondrial | Protein folding and maturation, response to unfolded protein | + | ||
Identification of carbonylated (+) proteins in non-AKU control (CTR), AKU “white” (CHW) and “black” (CHB) chondrocytes.
In control chondrocytes, we could identify as carbonylated only the proteins G3P (energy) and VIME (cell organization). In AKU cells, whereas “white” chondrocytes had no resolved immunoreactive spots but only high molecular weight protein aggregates stacked on the top of the SDS–PAGE gel, “black” AKU chondrocytes showed, together with carbonylated high molecular weight protein aggregates, the following carbonylated proteins: G3P (energy), EF1A1 (transcription, synthesis, and turnover of proteins), VIME (cell organization), CATA and CH60 (cell rescue, defence, and stress).
Discussion
In this study we have provided, to the best of our knowledge, the first proteomic and redox proteomic characterization of ex vivo AKU chondrocytes as models of ochronotic arthropathy. Our main aim was to gain insights into those AKU-related pathophysiological processes that allow ochronosis to develop in connective tissues, especially joints. We were able to subculture two distinct populations, namely “white” and “black” AKU chondrocytes, on the basis of the macroscopic appearance of donors' hip ochronotic cartilage from which the cells were obtained. Both cell types, maintained in culture conditions resembling the original pathological ones thanks to the addition of HGA to the culture medium, were characterized by increased apoptosis, NO release, and levels of pro-inflammatory cytokines similarly to what observed in OA, a pathology that share some features with alkaptonuric arthropathy. Interestingly, both the cell types contained intracellular ochronotic pigments, suggesting that ochronosis might involve joint cells before than the surrounding ECM. Profound alterations in the levels of proteins involved in cell defence, protein folding, and cell organization were found in AKU ochronotic chondrocytes together with an increased post-translational oxidation of proteins and oxidized high molecular weight protein aggregates.
One of the main functions of articular cartilage is to provide a structure dissipating excessive loading and withstanding tension, compression, and shearing forces (Juang et al., 2010). Chondrocytes are crucial players in maintaining cartilage integrity by activating catabolic or anabolic processes in response to external stimuli. The catabolic program activated upon inflammation involves the production of proteases and the induction of apoptosis together with the suppression of matrix synthesis. Conversely, the anabolic program promotes the secretion of specific cytokines, protease inhibitors, the production of ECM and cell proliferation (Lotz et al., 1995). An imbalance towards an increase in catabolic activities is thought to be involved in the disruption of cartilage integrity leading to loss of joint functions (Blanco Garcia, 1999; Kim and Blanco, 2007). We have already demonstrated an inflammatory status in AKU (Selvi et al., 2000) and IL-6 and IL-8 over-expression in AKU chondrocytes (unpublished data). In this work, we confirmed our previous findings and also highlighted that levels of NO were increased in AKU ochronotic cells concomitantly with an enhanced release of pro-inflammatory cytokines (IL-1β, IL-6, IL-8, and TNFα). Altogether, our results suggest that pro-inflammatory cytokines and NO might play important roles in cartilage and joint tissue disruption in AKU patients during the ochronotic progression.
NO plays pro-inflammatory and destructive effects in cartilages and it has been related to a variety of mechanisms promoting cartilage catabolism (Henrotin et al., 2005). Cytokines such as TNFα and IL-1β are important mediators of inflammation and cartilage degradation observed in OA (Cillero-Pastor et al., 2010). Both have been shown to modulate proteins regulating cytoskeleton, tissue organization, and cell morphology of human chondrocytes (Cillero-Pastor et al., 2010) and to induce the expression of PRDX1 (Cillero-Pastor et al., 2010), whose main function is to amend oxidative stress imposed by hydrogen peroxide. Similar results were confirmed in this work thanks to the proteomic characterization of AKU cells. TNFα was also found to down-regulate the expression of G3P and LDBH (Cillero-Pastor et al., 2010). Concomitantly with an enhanced release of TNFα, we observed in both AKU “white” and “black” chondrocytes a decrease in the abundance of G3P, whereas LDBH was under-expressed only in “white” chondrocytes. LDB is down-regulated also in OA patients (Ruiz-Romero et al., 2008). This suggests a lowering of the glycolysis rate and ATP production possibly predisposing to cell apoptosis, like observed upon TNFα stress (Cillero-Pastor et al., 2010), a finding corroborated in our work by a lower expression of ATPB protein (which is involved in ATP production) in “black” chondrocytes.
Inhibition of metabolism-related proteins is a common feature of OA (Iliopoulos et al., 2010). Nevertheless, the over-expression of proteins with functions in energy and protein fate, such as G3P, PDIA1, HS47, and CALR, as a function of catabolic program was highlighted in a rat chondrocyte model exposed to pressure overload (Juang et al., 2010). Interestingly, in our work we found that G3P, PDIA1, and CALR were under-expressed in AKU ochronotic chondrocytes, probably pointing out to an impaired ability of cells to produce a cartilage structure optimally withstanding pressure forces. In this light, it is noteworthy to mention that load bearing joints are the first to show the deposition of ochronotic pigments (La Du, 1991).
IL-1β is a key mediator of inflammation and cartilage degradation in OA, but it also play a role in oxidative stress generation since it can induce an imbalance in the activity of fundamental anti-oxidant enzymes, such as superoxide dismutase, catalase, and glutathione peroxidase, as demonstrated recently in chondrocytes (Mathy-Hartert et al., 2008). Though to a lesser extent, also IL-6 was found to induce such a disregulation of antioxidant enzymes. These changes can ultimately lead to a transient accumulation of ROS (Mathy-Hartert et al., 2008) which might induce, in turn, oxidation of cellular constituents. On this basis, and also taking into account the HGA-induced oxidative post-translational modifications of proteins that we highlighted in different AKU models (Braconi et al., 2010a, b; Tinti et al., 2010, 2011b), we performed a redox proteomic analysis to evaluate protein carbonylation in AKU ochronotic cells. Notably, in this work we found responses that were similar to what previously showed in a chondrocytic cell line treated with 0.33 mM HGA and used as an in vitro model of AKU ochronosis (Braconi et al., 2010b). We confirmed the identity of proteins most likely to undergo HGA-induced oxidation and also the production of high molecular weight aggregates of oxidized proteins, providing in turn indirect evidences of the auto-oxidation of HGA into BQA, which is known to promote protein aggregation mediated by oxidative phenomena. Protein oxidation and aggregation might help, in vivo, the production of ochronotic pigments.
In conclusion, the findings presented in this work suggest that alkaptonuric ochronosis might be mediated by profound alterations in chondrocyte structure and organization leading to an impaired ability of cartilages to withstand loading forces. Concomitantly, pro-inflammatory stimuli and NO-mediated damage can contribute to cartilage degradation and chondrocyte apoptosis. AKU ochronotic cells are also characterized by a lower expression of proteins for the protection against stresses (leading to consequent protein oxidation, induced by HGA and its conversion into BQA, a spontaneous reaction which is an additional source of ROS). This, together with a lower expression of proteins promoting protein folding, points towards an increased protein aggregation which might lay the basis in vivo for the production and deposition of ochronotic pigments.
Glossary
Abbreviations
- AKU
alkaptonuria
- BQA
benzoquinone acetic acid
- CHAPS
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate
- DNPH
2,4-dinitrophenylhydrazine
- DTE
1,4-dithioerythritol
- HGA
homogentisic acid
- IAA
iodoacetamide
- IEF
isoelectric focusing
- MS
mass spectrometry
- PAGE
polyacrylamide gel electrophoresis
- ROS
reactive oxygen species
- SDS
sodium dodecyl sulfate
Supplementary material
Literature Cited
- Aigner T, Kim HA. Apoptosis and cellular vitality: Issues in osteoarthritic cartilage degeneration. Arthritis Rheum. 2002;46:1986–1996. doi: 10.1002/art.10554. [DOI] [PubMed] [Google Scholar]
- Aizawa T, Kon T, Einhorn TA, Gerstenfeld LC. Induction of apoptosis in chondrocytes by tumor necrosis factor-alpha. J Orthop Res. 2001;19:785–796. doi: 10.1016/S0736-0266(00)00078-4. [DOI] [PubMed] [Google Scholar]
- Angeles AP, Badger R, Gruber HE, Seegmiller JE. Chondrocyte growth inhibition induced by homogentisic acid and its partial prevention with ascorbic acid. J Rheumatol. 1989;16:512–517. [PubMed] [Google Scholar]
- Blanco Garcia FJ. Catabolic events in osteoarthritic cartilage. Osteoarthritis Cartilage. 1999;7:308–309. doi: 10.1053/joca.1998.0174. [DOI] [PubMed] [Google Scholar]
- Braconi D, Laschi M, Amato L, Bernardini G, Millucci L, Marcolongo R, Cavallo G, Spreafico A, Santucci A. Evaluation of anti-oxidant treatments in an in vitro model of alkaptonuric ochronosis. Rheumatology (Oxford) 2010a;49:1975–1983. doi: 10.1093/rheumatology/keq175. [DOI] [PubMed] [Google Scholar]
- Braconi D, Laschi M, Taylor AM, Bernardini G, Spreafico A, Tinti L, Gallagher JA, Santucci A. Proteomic and redox-proteomic evaluation of homogentisic acid and ascorbic acid effects on human articular chondrocytes. J Cell Biochem. 2010b;111:922–932. doi: 10.1002/jcb.22780. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Capin-Gutierrez N, Talamas-Rohana P, Gonzalez-Robles A, Lavalle-Montalvo C, Kouri JB. Cytoskeleton disruption in chondrocytes from a rat osteoarthrosic (OA)-induced model: Its potential role in OA pathogenesis. Histol Histopathol. 2004;19:1125–1132. doi: 10.14670/HH-19.1125. [DOI] [PubMed] [Google Scholar]
- Cillero-Pastor B, Ruiz-Romero C, Carames B, Lopez-Armada MJ, Blanco FJ. Proteomic analysis by two-dimensional electrophoresis to identify the normal human chondrocyte proteome stimulated by tumor necrosis factor alpha and interleukin-1beta. Arthritis Rheum. 2010;62:802–814. doi: 10.1002/art.27265. [DOI] [PubMed] [Google Scholar]
- De Ceuninck F, Marcheteau E, Berger S, Caliez A, Dumont V, Raes M, Anract P, Leclerc G, Boutin JA, Ferry G. Assessment of some tools for the characterization of the human osteoarthritic cartilage proteome. J Biomol Tech. 2005;16:256–265. [PMC free article] [PubMed] [Google Scholar]
- Facchini A, Borzi RM, Marcu KB, Stefanelli C, Olivotto E, Goldring MB, Facchini A, Flamigni F. Polyamine depletion inhibits NF-kappaB binding to DNA and interleukin-8 production in human chondrocytes stimulated by tumor necrosis factor-alpha. J Cell Physiol. 2005;204:956–963. doi: 10.1002/jcp.20368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes JC, Martel-Pelletier J, Pelletier JP. The role of cytokines in osteoarthritis pathophysiology. Biorheology. 2002;39:237–246. [PubMed] [Google Scholar]
- Fernandez-Canon JM, Granadino B, Beltran-Valero deBernabe, Renedo D, Fernandez-Ruiz M, Penalva E, Rodriguez MA, de Cordoba S. The molecular basis of alkaptonuria. Nat Genet. 1996;14:19–24. doi: 10.1038/ng0996-19. [DOI] [PubMed] [Google Scholar]
- Fioravanti A, Nerucci F, Annefeld M, Collodel G, Marcolongo R. Morphological and cytoskeletal aspects of cultivated normal and osteoarthritic human articular chondrocytes after cyclical pressure: A pilot study. Clin Exp Rheumatol. 2003;21:739–746. [PubMed] [Google Scholar]
- Fischer BA, Mundle S, Cole AA. Tumor necrosis factor-alpha induced DNA cleavage in human articular chondrocytes may involve multiple endonucleolytic activities during apoptosis. Microsc Res Tech. 2000;50:236–242. doi: 10.1002/1097-0029(20000801)50:3<236::AID-JEMT7>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Flannery CR, Little CB, Hughes CE, Curtis CL, Caterson B, Jones SA. IL-6 and its soluble receptor augment aggrecanase-mediated proteoglycan catabolism in articular cartilage. Matrix Biol. 2000;19:549–553. doi: 10.1016/s0945-053x(00)00111-6. [DOI] [PubMed] [Google Scholar]
- Gobezie R, Kho A, Krastins B, Sarracino DA, Thornhill TS, Chase M, Millett PJ, Lee DM. High abundance synovial fluid proteome: Distinct profiles in health and osteoarthritis. Arthritis Res Ther. 2007;9:R36. doi: 10.1186/ar2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimmer C, Balbus N, Lang U, Aigner T, Cramer T, Muller L, Swoboda B, Pfander D. Regulation of type II collagen synthesis during osteoarthritis by prolyl-4-hydroxylases: Possible influence of low oxygen levels. Am J Pathol. 2006;169:491–502. doi: 10.2353/ajpath.2006.050738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell TR, Gallagher JA, Ranganath L. Alkaptonuria—A review of surgical and autopsy pathology. Histopathology. 2008;53:503–512. doi: 10.1111/j.1365-2559.2008.03000.x. [DOI] [PubMed] [Google Scholar]
- Henrotin Y, Kurz B, Aigner T. Oxygen and reactive oxygen species in cartilage degradation: Friends or foes? Osteoarthritis Cartilage. 2005;13:643–654. doi: 10.1016/j.joca.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Holloway I, Kayser M, Lee DA, Bader DL, Bentley G, Knight MM. Increased presence of cells with multiple elongated processes in osteoarthritic femoral head cartilage. Osteoarthritis Cartilage. 2004;12:17–24. doi: 10.1016/j.joca.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Honorati MC, Bovara M, Cattini L, Piacentini A, Facchini A. Contribution of interleukin 17 to human cartilage degradation and synovial inflammation in osteoarthritis. Osteoarthritis Cartilage. 2002;10:799–807. doi: 10.1053/joca.2002.0829. [DOI] [PubMed] [Google Scholar]
- Iliopoulos D, Gkretsi V, Tsezou A. Proteomics of osteoarthritic chondrocytes and cartilage. Expert Rev Proteomics. 2010;7:749–760. doi: 10.1586/epr.10.27. [DOI] [PubMed] [Google Scholar]
- Isomaki P, Punnonen J. Pro- and anti-inflammatory cytokines in rheumatoid arthritis. Ann Med. 1997;29:499–507. doi: 10.3109/07853899709007474. [DOI] [PubMed] [Google Scholar]
- Jorgensen C, Apparailly F, Couret I, Canovas F, Jacquet C, Sany J. Interleukin-4 and interleukin-10 are chondroprotective and decrease mononuclear cell recruitment in human rheumatoid synovium in vivo. Immunology. 1998;93:518–523. doi: 10.1046/j.1365-2567.1998.00457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juang YM, Lee CY, Hsu WY, Lin CT, Lai CC, Tsai FJ. Proteomic analysis of chondrocytes exposed to pressure. Biomed Chromatogr. 2010;24:1273–1282. doi: 10.1002/bmc.1436. [DOI] [PubMed] [Google Scholar]
- Katsikis PD, Chu CQ, Brennan FM, Maini RN, Feldmann M. Immunoregulatory role of interleukin 10 in rheumatoid arthritis. J Exp Med. 1994;179:1517–1527. doi: 10.1084/jem.179.5.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HA, Blanco FJ. Cell death and apoptosis in osteoarthritic cartilage. Curr Drug Targets. 2007;8:333–345. doi: 10.2174/138945007779940025. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick CJ, Mohr W, Mutschler W. Experimental studies on the pathogenesis of ochronotic arthropathy. The effects of homogentisic acid on adult and fetal articular chondrocyte morphology, proliferative capacity and synthesis of proteoglycans in vitro. Virchows Arch B Cell Pathol Incl Mol Pathol. 1984;47:347–360. [PubMed] [Google Scholar]
- Kobayashi M, Squires GR, Mousa A, Tanzer M, Zukor DJ, Antoniou J, Feige U, Poole AR. Role of interleukin-1 and tumor necrosis factor alpha in matrix degradation of human osteoarthritic cartilage. Arthritis Rheum. 2005;52:128–135. doi: 10.1002/art.20776. [DOI] [PubMed] [Google Scholar]
- Kokebie R, Aggarwal R, Lidder S, Hakimiyan AA, Rueger DC, Block JA, Chubinskaya S. The role of synovial fluid markers of catabolism and anabolism in osteoarthritis, rheumatoid arthritis and asymptomatic organ donors. Arthritis Res Ther. 2011;13:R50. doi: 10.1186/ar3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutty MK, Iqbal QM, Teh EC. Ochronotic arthropathy. A comparative study of light microscopic and ultrastructural features. Arch Pathol. 1973;96:100–103. [PubMed] [Google Scholar]
- La Du BN., Jr Alcaptonuria and ochronotic arthritis. Mol Biol Med. 1991;8:31–38. [PubMed] [Google Scholar]
- La Du BN, Zannoni VG, Laster L, Seegmiller JE. The nature of the defect in tyrosine metabolism in alcaptonuria. J Biol Chem. 1958;230:251–260. [PubMed] [Google Scholar]
- Lechman ER, Jaffurs D, Ghivizzani SC, Gambotto A, Kovesdi I, Mi Z, Evans CH, Robbins PD. Direct adenoviral gene transfer of viral IL-10 to rabbit knees with experimental arthritis ameliorates disease in both injected and contralateral control knees. J Immunol. 1999;163:2202–2208. [PubMed] [Google Scholar]
- Li H, Yang HS, Wu TJ, Zhang XY, Jiang WH, Ma QL, Chen YX, Xu Y, Li S, Hua ZC. Proteomic analysis of early-response to mechanical stress in neonatal rat mandibular condylar chondrocytes. J Cell Physiol. 2010;223:610–622. doi: 10.1002/jcp.22052. [DOI] [PubMed] [Google Scholar]
- Lotz M, Blanco FJ, von Kempis J, Dudler J, Maier R, Villiger PM, Geng Y. Cytokine regulation of chondrocyte functions. J Rheumatol Suppl. 1995;43:104–108. [PubMed] [Google Scholar]
- Mathy-Hartert M, Hogge L, Sanchez C, Deby-Dupont G, Crielaard JM, Henrotin Y. Interleukin-1beta and interleukin-6 disturb the antioxidant enzyme system in bovine chondrocytes: A possible explanation for oxidative stress generation. Osteoarthritis Cartilage. 2008;16:756–763. doi: 10.1016/j.joca.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Moo V, Sieper J, Herzog V, Muller BM. Regulation of expression of cytokines and growth factors in osteoarthritic cartilage explants. Clin Rheumatol. 2001;20:353–358. doi: 10.1007/s100670170025. [DOI] [PubMed] [Google Scholar]
- Muller RD, John T, Kohl B, Oberholzer A, Gust T, Hostmann A, Hellmuth M, Laface D, Hutchins B, Laube G, Veh RW, Tschoeke SK, Ertel W, Schulze-Tanzil G. IL-10 overexpression differentially affects cartilage matrix gene expression in response to TNF-alpha in human articular chondrocytes in vitro. Cytokine. 2008;44:377–385. doi: 10.1016/j.cyto.2008.10.012. [DOI] [PubMed] [Google Scholar]
- O'Brien WM, La Du BN, Bunim JJ. Biochemical, pathologic and clinical aspects of alkaptonuria, ochronosis and ochronotic arthropathy: Review of world literature (1584–1962) Am J Med. 1963;34:813–838. [Google Scholar]
- Pelletier JP, Martel-Pelletier J, Abramson SB. Osteoarthritis, an inflammatory disease: Potential implication for the selection of new therapeutic targets. Arthritis Rheum. 2001;44:1237–1247. doi: 10.1002/1529-0131(200106)44:6<1237::AID-ART214>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Phornphutkul C, Introne WJ, Perry MB, Bernardini I, Murphey MD, Fitzpatrick DL, Anderson PD, Huizing M, Anikster Y, Gerber LH, Gahl WA. Natural history of alkaptonuria. N Engl J Med. 2002;347:2111–2121. doi: 10.1056/NEJMoa021736. [DOI] [PubMed] [Google Scholar]
- Rediske JJ, Koehne CF, Zhang B, Lotz M. The inducible production of nitric oxide by articular cell types. Osteoarthritis Cartilage. 1994;2:199–206. doi: 10.1016/s1063-4584(05)80069-x. [DOI] [PubMed] [Google Scholar]
- Reinheckel T, Korn S, Mohring S, Augustin W, Halangk W, Schild L. Adaptation of protein carbonyl detection to the requirements of proteome analysis demonstrated for hypoxia/reoxygenation in isolated rat liver mitochondria. Arch Biochem Biophys. 2000;376:59–65. doi: 10.1006/abbi.1999.1680. [DOI] [PubMed] [Google Scholar]
- Ruiz-Romero C, Carreira V, Rego I, Remeseiro S, Lopez-Armada MJ, Blanco FJ. Proteomic analysis of human osteoarthritic chondrocytes reveals protein changes in stress and glycolysis. Proteomics. 2008;8:495–507. doi: 10.1002/pmic.200700249. [DOI] [PubMed] [Google Scholar]
- Selvi E, Manganelli S, Mannoni A, Benucci M, Minacci C, Marcolongo R. Chronic ochronotic arthritis: Clinical, arthroscopic, and pathologic findings. J Rheumatol. 2000;27:2272–2274. [PubMed] [Google Scholar]
- Spreafico A, Frediani B, Capperucci C, Chellini F, Paffetti A, D'Ambrosio C, Bernardini G, Mini R, Collodel G, Scaloni A, Marcolongo R, Santucci A. A proteomic study on human osteoblastic cells proliferation and differentiation. Proteomics. 2006;6:3520–3532. doi: 10.1002/pmic.200500858. [DOI] [PubMed] [Google Scholar]
- Spreafico A, Chellini F, Frediani B, Bernardini G, Niccolini S, Serchi T, Collodel G, Paffetti A, Fossombroni V, Galeazzi M, Marcolongo R, Santucci A. Biochemical investigation of the effects of human platelet releasates on human articular chondrocytes. J Cell Biochem. 2009;108:1153–1165. doi: 10.1002/jcb.22344. [DOI] [PubMed] [Google Scholar]
- Switzer RC, III, Merril CR, Shifrin S. A highly sensitive silver stain for detecting proteins and peptides in polyacrylamide gels. Anal Biochem. 1979;98:231–237. doi: 10.1016/0003-2697(79)90732-2. [DOI] [PubMed] [Google Scholar]
- Theos AC, Tenza D, Martina JA, Hurbain I, Peden AA, Sviderskaya EV, Stewart A, Robinson MS, Bennett DC, Cutler DF, Bonifacino JS, Marks MS, Raposo G. Functions of adaptor protein (AP)-3 and AP-1 in tyrosinase sorting from endosomes to melanosomes. Mol Biol Cell. 2005;16:5356–5372. doi: 10.1091/mbc.E05-07-0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinti L, Spreafico A, Braconi D, Millucci L, Bernardini G, Chellini F, Cavallo G, Selvi E, Galeazzi M, Marcolongo R, Gallagher JA, Santucci A. Evaluation of antioxidant drugs for the treatment of ochronotic alkaptonuria in an in vitro human cell model. J Cell Physiol. 2010;225:84–91. doi: 10.1002/jcp.22199. [DOI] [PubMed] [Google Scholar]
- Tinti L, Spreafico A, Chellini F, Galeazzi M, Santucci A. A novel ex vivo organotypic culture model of alkaptonuria-ochronosis. Clin Exp Rheumatol. 2011a;29:693–696. [PubMed] [Google Scholar]
- Tinti L, Taylor AM, Santucci A, Wlodarski B, Wilson PJ, Jarvis JC, Fraser WD, Davidson JS, Ranganath LR, Gallagher JA. Development of an in vitro model to investigate joint ochronosis in alkaptonuria. Rheumatology. 2011b;50:271–277. doi: 10.1093/rheumatology/keq246. [DOI] [PubMed] [Google Scholar]
- Umlauf D, Frank S, Pap T, Bertrand J. Cartilage biology, pathology, and repair. Cell Mol Life Sci. 2010;67:4197–4211. doi: 10.1007/s00018-010-0498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehling N, Palmer GD, Pilapil C, Liu F, Wells JW, Muller PE, Evans CH, Porter RM. Interleukin-1beta and tumor necrosis factor alpha inhibit chondrogenesis by human mesenchymal stem cells through NF-kappaB-dependent pathways. Arthritis Rheum. 2009;60:801–812. doi: 10.1002/art.24352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannoni VG, Malwista SE, La Du BN. Studies on ochronosis II. Studies on benzoquinoneacetic acid, a probable intermediate in the connective tissue pigmentation in alkaptonuria. Arthritis Rheum. 1962;5:547–556. doi: 10.1002/art.1780050603. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


