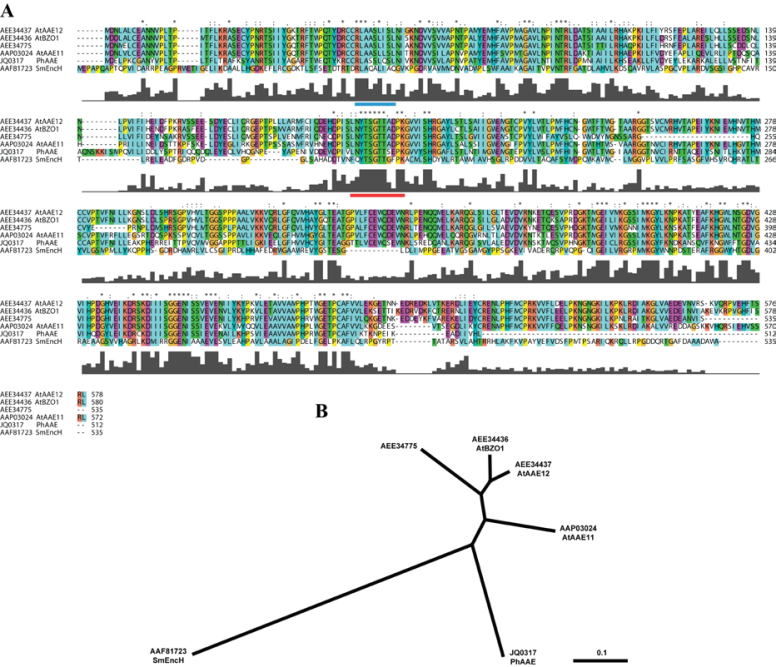
Fig. 2.
Predicted amino acid sequence alignment and phylogenetic tree. Sequences represented are from Streptomyces maritimus, Arabidopsis thaliana, and Petunia×hybrida. Sequences were aligned using MULTIPLE ALIGNMENT MODE of ClustalX 2.0 program software (Larkin et al., 2007). Above the sequence alignment ‘*’ indicates a fully conserved residue, ‘:’ indicates a fully conserved ‘strong’ group, and ‘.’ indicates a fully conserved ‘weak’ group. Colours are assigned to groups of amino acids as follows: orange for G, P, S, and T; red for H, K, and R; blue for F, W, and Y; and green for I, L, M, and V. Predicted PTS2 (Do-It-Yourself Block Search http://www.peroxisomedb.org/diy_PTS1.html) is represented by a blue bar below residues 51–59 of PhAAE, while the conserved AMP-binding site (InterProScan, http://www.ebi.ac.uk/Tools/pfa/iprscan/) is represented by a red bar below residues 197–208 of PhAAE (A). The Neighbor–Joining tree was derived by ClustalX and visualized in TreeView 1.6.6 as an unrooted tree (B).