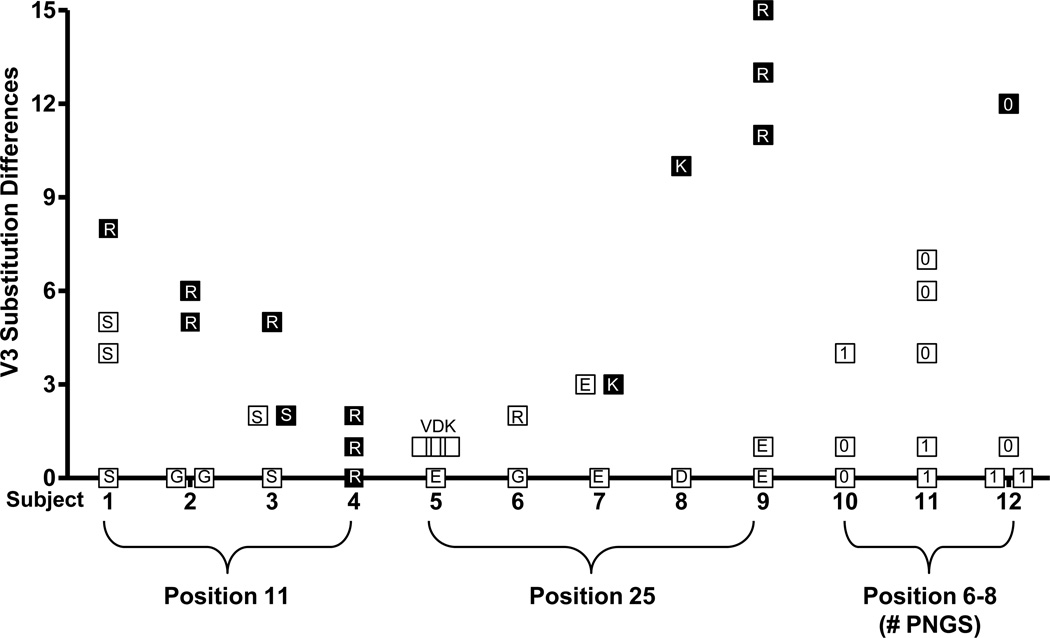
Figure 1. The number of V3 amino acid differences among env clones from each of 12 subjects.
Each symbol represents a unique V3 sequences that differs from the most prevalent V3 sequence of R5 clones in the same sample (except for subject 4). Subject 4 contained only dual-X clones (efficiently utilize CXCR4) and the number of amino acid differences between clones with or without 11R is shown. R5 and dual-R (inefficiently utilize CXCR4) clones are indicated by open squares, dual-X and X4 clones are indicated by filled squares. The amino acid substitution or number of PNGS is shown within each symbol.