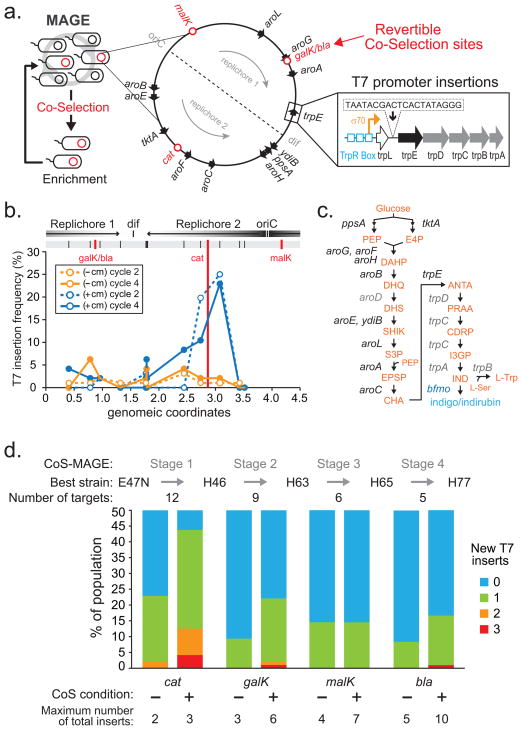
Fig 1. Co-selection MAGE strategy for enriching highly modified genomes.
a, Cells undergoing cycles of MAGE were enriched by a co-selection stage through phenotypic selection of a revertible genomic locus (i.e. malK, galK, cat or bla) and used for subsequent cycles of MAGE in an iterative fashion. Twelve genomic operons were targeted for insertion of the T7 promoter sequence upstream of the first relevant open reading frame (ORF). b, Enhanced T7 insertion frequency near the cat locus was observed only when the population was co-selected by chloramphenicol for cat (blue lines) in comparison to without co-selection (orange lines) after two (dotted lines) and four (solid lines) MAGE cycles as determined by MASC-PCR (Online Methods). c, Biosynthesis of tryptophan and indole derivatives indigo and indirubin. Intermediate products are shown in orange and the first gene in each relevant operon shown in black. Targeted genes that are not first in the operon are shown in gray. d, Each stage of CoS-MAGE involves 4 cycles of MAGE followed by screening of 96 colonies to find the most modified mutant. Population frequency of colonies with 0 to 3 new T7p insertions for each co-selection stage are shown for the corresponding conditions. Co-selection enriches for highly modified genomes, which are used in the subsequent CoS-MAGE stages with a reduced oligo pool to target the remaining sites. The most enriched strains at the end of each CoS stage are noted (H46, H63, H65, H77).