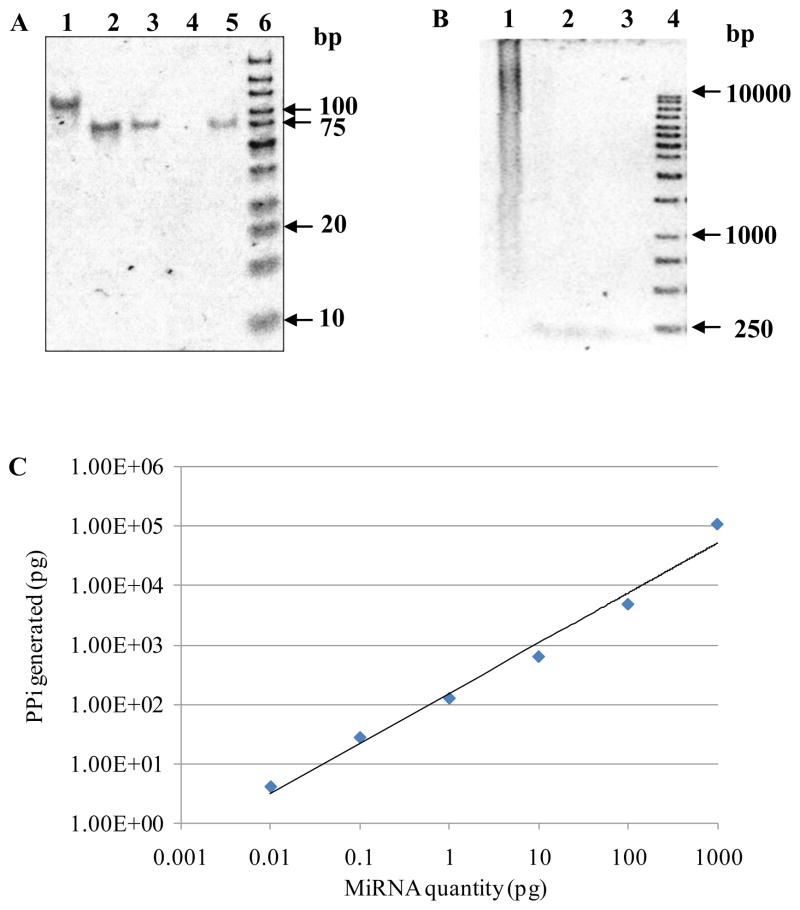
Figure 3.
Sensitivity and dynamic range of the enzymatic luminescence assay. (A) 15% acrylamide/7M urea denaturing gel electrophoresis of closed circular ssDNA converted from a linear ssDNA by CircLigase. Lane 1: closed circular ssDNA reaction product. Lane 2: linear oligonucleotide probe hsa-miR-26a, 50 ng. Lane 3: linear oligonucleotide probe hsa-miR-26a, 10 ng. Lane 4: blank. Lane 5: 71-base-long linear oligonucleotide probe, 8 ng. Lane 6: O’GeneRuler™ ultra low range DNA ladder, 1 μl. (B) 1% agarose gel electrophoresis of RCA reaction. Lane 1: RCA reaction products from synthetic circular ssDNA and 1 ng complementary RNA target. Lane 2: circular ssDNA. Lane 3: duplex of circular ssDNA and its complementary RNA target. Lane 4: O’GeneRuler™ 1 kb DNA ladder. (C) Sensitivity and dynamic range of the enzymatic luminescence assay for miRNA detection.