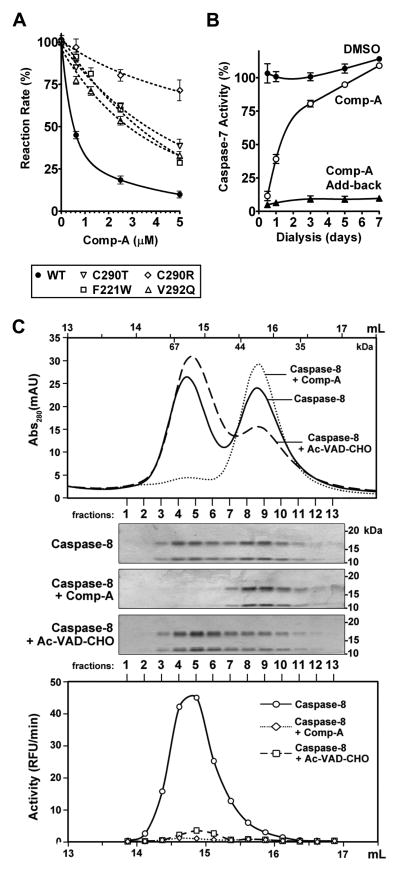
Figure 7. Comp-A Inhibits Effective Dimerization of Caspases in a Noncovalent, Reversible Manner.
(A) Mutation of C290, F221, and V292 affect caspase-7 inhibition by Comp-A. Initial reaction rates of wild-type and mutant caspase-7 were measured in the presence of Comp-A at the indicated concentrations. Reaction rates are expressed relative to the DMSO control. Error bars represent the SEM from triplicate experiments.
(B) Inhibition of caspase-7 by Comp-A is reversible. 200 nM of caspase-7 were incubated with either DMSO (labeled “DMSO”) or 10 μM Comp-A (labeled “Comp-A”). The samples were dialyzed against buffer ASC. Aliquots were removed at the indicated time-points and caspase-7 activity was measured with or without the addition of 10 μM of exogenous Comp-A (labeled “Comp-A Add-back”). Activity is expressed relative to un-dialyzed Caspase-7. Error bars represent the SEM from triplicate experiments.
(C) Comp-A disrupts dimerization of caspase-8. 100 nM of recombinant caspase-8 were incubated with DMSO, 10 μM Comp-A, or 10 μM Ac-VAD-CHO for 15 minutes at 30°C and then subjected to Superdex 200 gel filtration. Collected fractions were subjected to SDS-PAGE stained with Coomassie Blue (middle). Activity in each fraction was measured (bottom plot). The volume (mL) labels on the top and bottom plots indicate the elution volume of the chromatographic run (starting from sample injection). The elution positions of protein mass standards are labeled inside of the top Abs280 plot.