Figure 1.
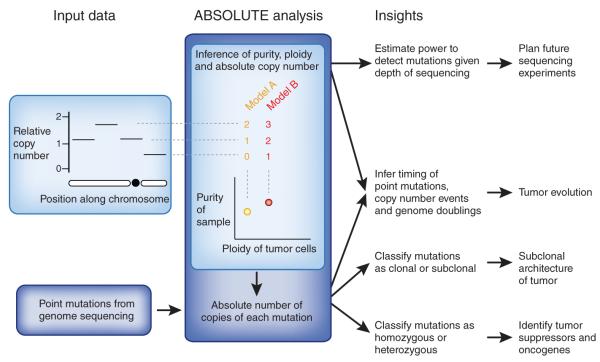
Biological insights obtained from sophisticated methods for copy number inference. From SNP array or high-throughput sequencing data, methods such as ABSOLUTE can infer tumor purity and ploidy and genome-wide, absolute copy-number profiles. These methods leverage mathematical relationships between purity, ploidy and copy number at a locus and across the genome. Two alternative models are shown, corresponding to different combinations of purity and ploidy, for assignment of relative copy number to absolute copy number states. Purity and ploidy can be used to estimate the required depth of coverage to detect (clonal or subclonal) mutations with a given power. This is useful for designing subsequent sequencing experiments. Integrating copy number data with point mutation data makes it possible to calculate the absolute number of copies of each mutation in the genome. Tumor evolution can be studied by deducing the relative order in which point mutations, copy number events and genome doublings arose in the cancer genome. Separation of clonal from subclonal mutations provides insight into the subclonal architecture of tumors. Finally, homozygous mutations can be distinguished from heterozygous ones, facilitating the identification of tumor suppressors and oncogenes, respectively.