Figure S5.
Timing of Chromosomal Gains and Genomic Amplifications, Related to Figure 4
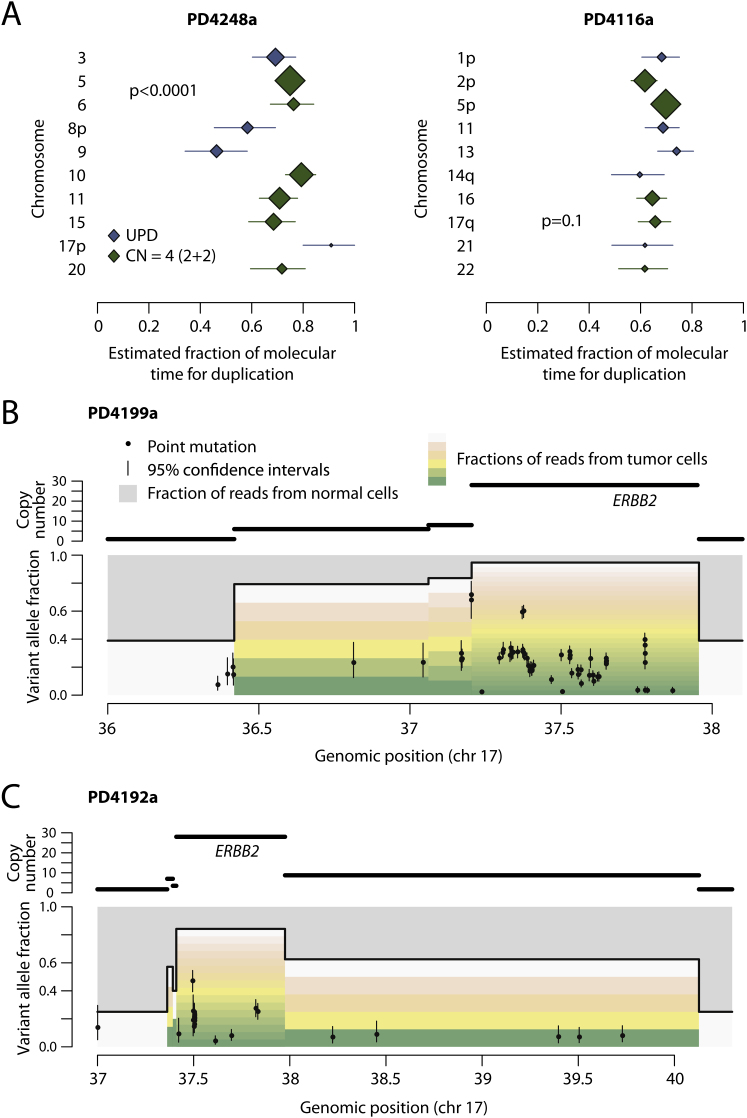
(A) Forest plots showing the point estimates (diamonds) and 95% confidence intervals estimated by bootstrapping of particular chromosomes for two breast cancer genomes. The size of the diamond is proportional to the number of mutations considered, and the color by whether the chromosomal gain reflects uniparental disomy (blue) or tetraploid chromosomes (green). The estimates show significant heterogeneity for PD4248a (p < 0.0001) but not for PD4116a (p = 0.3), with the latter indicating the possibility of all the gains occurring as a single endoreduplication event.
(B and C) Timing of ERBB2 genomic amplification for PD4199a (B) and PD4192a (C). Here, the top panel shows the copy number segments for the region of chromosome 17 around ERBB2. The lower panel shows the point mutations as black points, with the x axis reflecting the genomic position and the y axis the variant allele fraction. The 95% confidence intervals for the variant allele fraction are shown as vertical bars for each mutation. The allele fraction is divided by the colored bars into the proportions of reads derived from contaminating normal cells (gray bars) and the fraction coming from each of the copies of that segment in the tumor cells (the multiple bars from green to yellow to pink to white). Early mutations will be found relatively higher up these bars, whereas late ones will be seen toward the bottom of the variant allele fraction.