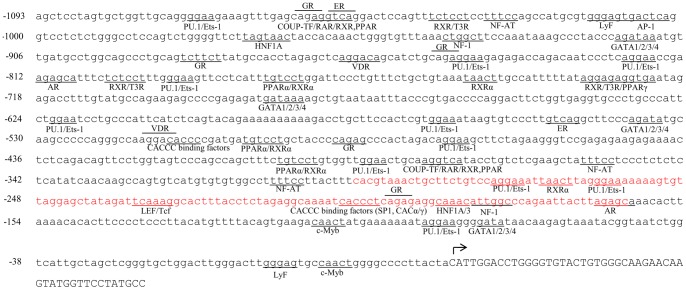
Figure 3. In silico analysis of mouse PXR proximal promoter.
The nucleotide sequence of the 5′ flanking genomic DNA of the mouse PXR gene is shown along with consensus sequences for potential DNA-protein binding sites. Transcription initiation site, +1, is denoted by an arrow. PU.1, purine rich box binding element; Ets-1, Ets binding element; GR, glucocorticoid receptor binding element; ER, estrogen receptor binding element; VDR, vitamin D receptor binding element; COUP-TF, chicken ovalbumin upstream promoter binding element; NF-AT, nuclear factor of activated T cells binding element; AP-1, activator protein binding element; HNF1/3/4, hepatocyte nuclear factor binding element; AR, androgen receptor binding element; RXR/RAR, retinoid X/acid receptor binding site; T3R, thyroid hormone receptor binding element; PPAR, peroxisome proliferator-activated receptor binding element; GATA, gata binding element; LEF, lymphoid enhancer factor binding element; LyF, lymphoid factor binding element; c-Myb, myb (myeloblast) binding element; NF-1, nuclear factor binding element. Region marked with red color shows putative binding sites for different transcription factors and this region is further used for EMSA analysis.