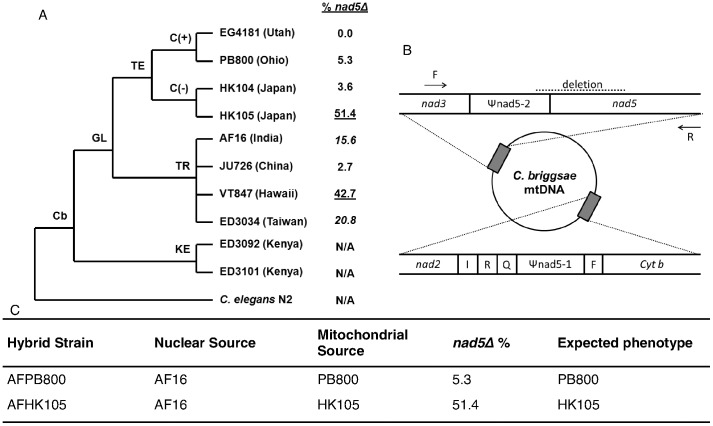
Figure 1. Natural and experimental C. briggsae strains and description of the nad5Δ mtDNA deletion.
A. Phylogenetic relationship and nad5Δ heteroplasmy level of C. briggsae isolates studied here. GL = global superclade; KE = Kenya clade; TE and TR = temperate and tropical subclades of GL; C(+) = isolates bearing compensatory Ψnad5Δ-2 allele; C(-) = isolates bearing ancestral alleles. nad5Δ heteroplasmy categories were assigned to each C. briggsae natural isolate for statistical analysis following Estes et al. (2011): High = underlined font, medium = italicized, low = regular, and zero-nad5Δ="N/A”. Note that we assayed the natural HK104 isolate here instead of the mutation-accumulation line progenitor reported in Estes et al. (2011), which had evolved high nad5Δ levels in the lab (See Materials and Methods). B. Positions of the nad5Δ deletion (dashed line at top) and Ψnad5Δ-2 elements in the mitochondrial genome. Primers are indicated by arrows (adapted from Howe and Denver, 2008). C. Mitochondrial and nuclear parent isolates for each mitochondrial-nuclear hybrid. nad5Δ heteroplasmy for each hybrid strain matches that of the maternal isolates as expected. Mitochondrial phenotypes are expected to match those of the maternal isolate if measured traits are predominantly determined by the mitochondrial genotype.