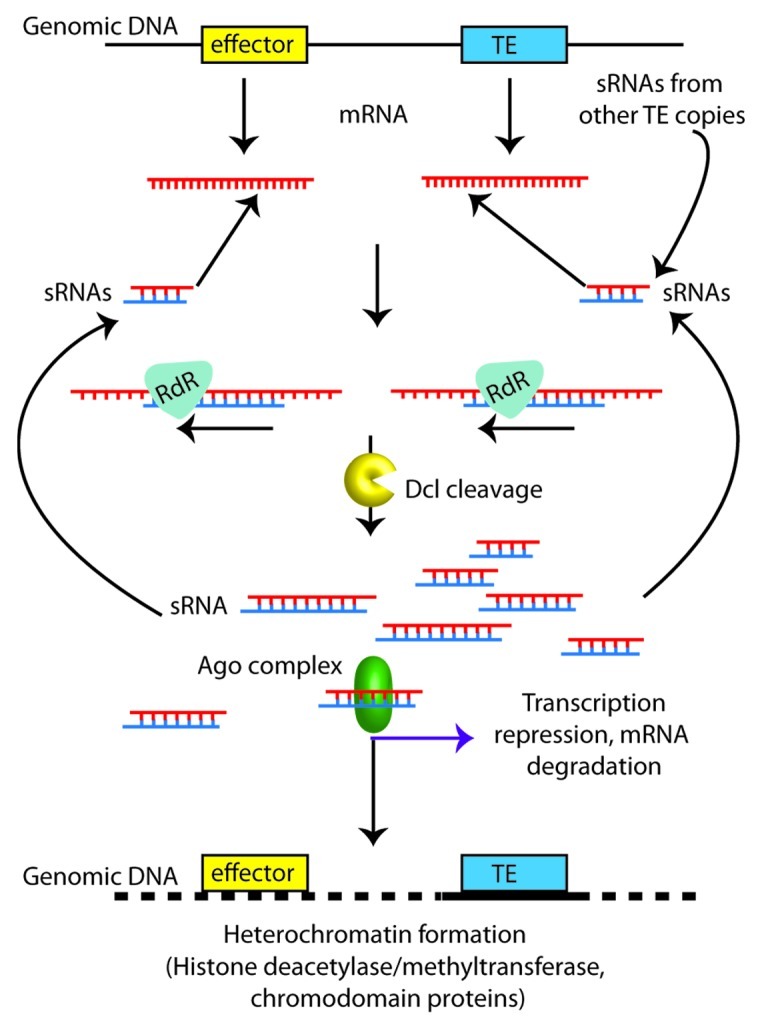
Figure 2. Proposed model for transcriptional repression of effector genes due to proximity to transposon sequences. Transposon sequences (TE; blue box) are strongly targeted for silencing by abundant homologous small RNAs (sRNAs). Inactivation of transposons results from formation of heterochromatin, guided by sRNAs. The heterochromatic region may spread outwards along the genomic DNA (dotted line), and affect nearby effector gene sequences (yellow), either through degradation of mRNAs (Ago) or repression of transcription (histone methylation). The formation of additional sRNAs, maintaining or reinforcing the silenced state, can occur through the action of Rdr and Dcl on mRNAs from the transposon or effector gene.
