Abstract
Conjugation is certainly the most widespread and promiscuous mechanism of horizontal gene transfer in bacteria. During conjugation, DNA translocation across membranes of two cells forming a mating pair is mediated by two types of mobile genetic elements: conjugative plasmids and integrating conjugative elements (ICEs). The vast majority of conjugative plasmids and ICEs employ a sophisticated protein secretion apparatus called type IV secretion system to transfer to a recipient cell. Yet another type of conjugative DNA translocation machinery exists and to date appears to be unique to conjugative plasmids and ICEs of the Actinomycetales order, a sub-group of high G + C Gram-positive bacteria. This conjugative system is reminiscent of the machinery that allows segregation of chromosomal DNA during bacterial cell division and sporulation, and relies on a single FtsK-homolog protein to translocate double-stranded DNA molecules to the recipient cell. Recent thorough sequence analyses reveal that while this latter strategy appears to be used by the majority of ICEs in Actinomycetales, the former is also predicted to be important in exchange of genetic material in actinobacteria.
Keywords: actinobacteria, integrating conjugative elements, conjugation, DNA translocation, horizontal gene transfer
Integrating Conjugative Elements
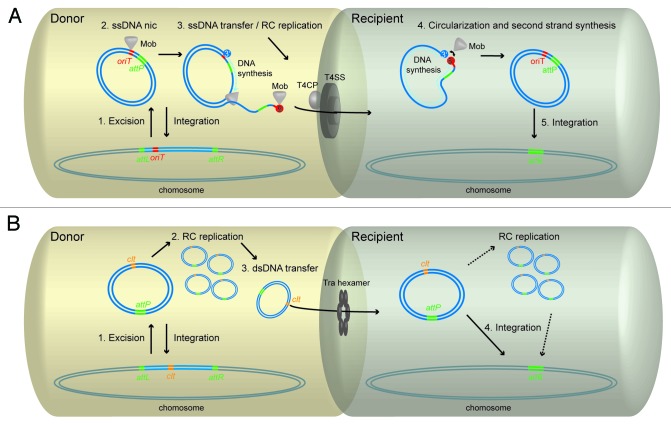
Conjugative DNA transfer allows rapid adaptation of bacteria through leaps of acquisition and exchange of massive amounts of genetic material even between distantly related microorganisms. While conjugative plasmids maintain in the host genome by autonomous replication, integrating conjugative elements (ICEs) have the ability to integrate within the host’s chromosome to be vertically inherited (for reviews see refs. 1 and 2). Consequently, ICEs need to excise from a donor cell’s chromosome into a circular form prior to transfer (Fig. 1). Integration and excision of ICEs are recombination events catalyzed by serine or tyrosine integrases (Int) between short homologous sequences called attachment sites (att), on the circular element (attP) and the chromosome (attB), or flanking the integrated element (attL and attR), respectively (Fig. 1). Although they share the same preliminary step, the mechanisms of conjugative transfer of ICEs and actinomycete ICEs (AICEs) fundamentally differ. Conjugative transfer of ICEs is presumed to be mechanistically similar to conjugative transfer of prototypical Gram-negative bacteria conjugative plasmids, while the mechanism of AICEs transfer is rather reminiscent of the one used by Streptomyces conjugative plasmids.3,4
Figure 1. Conjugative transfer models of ICEs from the two superfamilies. (A): (1) In the donor cell, ICE excision from the chromosome results from site-specific recombination between the attL and attR sites. Following excision, the relaxase (Mob), which is part of a multiprotein complex called relaxosome, recognizes the origin of transfer (oriT). (2) The Mob protein generates a nick in one strand and becomes covalently bound to the 5′ end of the nicked strand. (3) While the single-stranded nucleoprotein complex is displaced by ongoing rolling-circle (RC) replication, it interacts with the type IV coupling protein (T4CP) which generates the energy for its translocation through a dedicated type IV secretion system (T4SS). (4) Once transferred in the recipient cell, the Mob protein ligates the single-stranded DNA molecule and the complementary strand is synthesized. (5) Integration in the recipient cell’s chromosome is mediated by recombination between the attP site on the circular ICE and the chromosomal attB site. (B): (1) Like ICEs, AICEs excise from the chromosome by site-specific recombination. (2) The excised circular AICE then replicates by RC replication and reintegrate into the chromosome and/or transfer to a recipient cell by conjugation. (3) The transfer protein Tra recognizes the AICE cis-acting locus (clt) and mediates the transfer of the double-stranded AICE by forming a pore (Tra hexamer) in the lipid bilayer and the use of its ATPase activity. (4) The circular AICE integrates into the chromosome by site-specific recombination as described above. Alternatively, integration into the chromosome of the recipient cell could be preceded by an additional step in which RC replication would occur.
T4SS-Mediated Translocation of Single-Stranded DNA (ICEs)
Widespread in Gram-negative bacteria, conjugation mediated by type IV secretion systems (T4SS) requires the assembly of the mating pore, which varies in complexity, and usually involves secretion and assembly of an extracellular pilus.5,6 One of the key components of the T4SS is a VirB4-like sub-unit, which exhibits ATPase activity and likely energizes the assembly and/or activity of the secretion channel. Biochemical processing of the DNA molecule to transfer is initiated at the origin of transfer (oriT), which is bound by a DNA relaxase (Mob protein) and other auxiliary proteins (Fig. 1A). Altogether they assemble as a nucleoprotein complex, the relaxosome, which is recognized as a T4SS substrate.3,5,7 The phosphodiesterase activity of the relaxase mediates a strand-specific cleavage within oriT, allowing the unwinding of the DNA molecule and 5′ to 3′ transfer of a single-stranded DNA to the recipient cell. Another key component found associated with most conjugative T4SS is the coupling protein (T4CP), a VirD4-like subunit, which likely acts as a docking site for T4SS substrates. T4CPs are phylogenetically and structurally related to FtsK and SpoIIIE ATPases and power translocation of single-stranded DNA across the donor and recipient cell membranes.
TraB-Mediated Translocation of Double-Stranded DNA (AICEs)
FtsK-homolog based conjugative DNA-translocation systems are structurally simpler, relying on a single protein, TraB, aka TraSA or Tra, which resembles the septal DNA translocator FtsK.4 FtsK mediates proper segregation of the freshly duplicated circular chromosomal DNA into daughter-cell compartments during constriction of the septal membranes in prokaryotic cell division.8-10 AICEs likely transfer following the mechanism recently demonstrated for pSVH1 from Streptomyces venezuelae (Fig. 1B).4 Like for pSVH1, TraB-homologs encoded by AICEs would recognize and bind to a specific double-stranded DNA region on the circularized AICE, the cis-acting locus of transfer (clt) which is conceptually equivalent to the oriT of ICEs and necessary for efficient transfer. TraB of pSVH1 has been shown to oligomerise, forming a hexameric pore structure that is large enough to translocate double-stranded DNA. Since translocation of double-stranded DNA is not a conservative mechanism, transfer to the recipient of a mobile genetic element (MGE) using this strategy would ultimately lead to its loss from the donor cell. To circumvent this limitation, a dedicated rolling-circle replication module (Rep) mediates replication of AICEs after excision from the chromosome and prior to transfer to the recipient.11
Prevalence of AICEs
One of the earliest genome-wide identification of AICEs was performed by te Poele et al. by using sequence homology-based methods on actinomycete genomes.12 Recently, the prevalence of AICEs in 275 chromosomes and 176 plasmids of actinobacteria was estimated using methods based on hidden Markov model (HMM) protein profiles to search for various proteins families involved in maintenance [serine and tyrosine recombinases (Int), replication initiator proteins (Rep)] and transfer [FtsK-like conjugative DNA-translocation proteins (Tra)] of AICEs.13 These extensive in silico analyses revealed 144 putative AICEs using an FtsK-like Tra protein. With one exception found in a Bifidobacterium strain, all of these AICEs are exclusively detected in genomes of members of the Actinomycetales order (Fig. 2). The apparent absence of AICEs in the other actinobacteria subclasses (Acidimicrobidae, Coriobacteridae and Rubrobacteridae) is intriguing and tends to justify their classification as AICEs. The quasi-absence of FtsK-like Tra-based conjugative mobile elements in the genomes of non-Actinomycetales actinobacteria suggests that this mechanism of transfer is specifically adapted to the hyphal nature of the Actinomycetales. On the opposite, conjugative elements using this strategy may be unable to efficiently spread and therefore persist in populations of actinobacteria species growing as cocci or short chains.
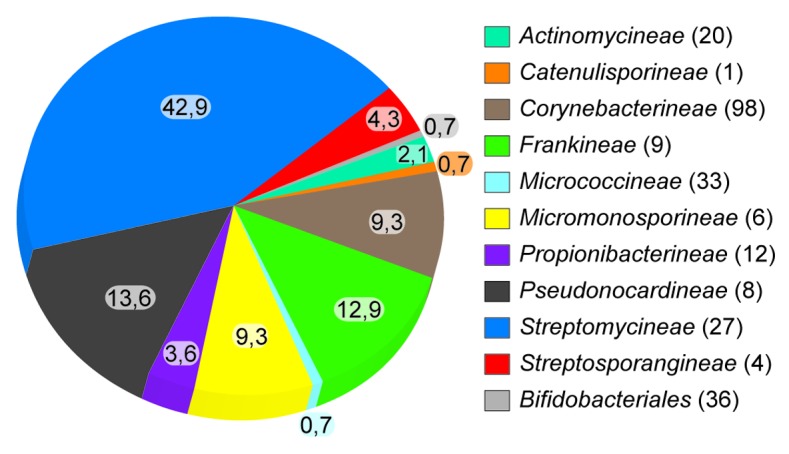
Figure 2. Taxonomic distribution of AICEs. Percent of the 140 AICEs detected in complete (130) and draft (145) actinobacterial genomes is indicated for each clade. Numbers in parentheses represent the number of genomes analyzed for each clade. The clades for which no AICEs could be found are not shown [Acidimicrobidae (1) Coriobacteridae (15), Rubrobacteridae (2), Glycomycineae (1), Kineosporiineae (1), unclassified actinobacteria (1)]. The four AICEs detected in the analysis of 176 actinobacterial plasmids are not included.
Single genomes hosting multiple different AICEs are frequent and the occurrence of AICEs correlates rather well with genome size. The genome of members of the Frankinaea, Micromonosporineae and Streptomycineae sub-orders seem to be more prone to harbor multiple unrelated AICEs, particularly in species isolated from plants, soil and water.13 This observation suggests that these specific niches likely favor cell-to-cell contacts but also provide frequent opportunities of contact between unrelated or distantly related bacterial partners. Conversely, genomes of bacteria isolated from dairy products, animals, human or insects rarely harbor multiple AICEs, likely reflecting the specialization of these more insulated, less diverse microbial floras.
AICEs Canonical Proteins
Analysis of the relationships between the putative Int, Rep and FtsK-like Tra proteins encoded by all 144 predicted AICEs, together with proteins of other mobile genetic elements, reveals for the first time the considerable diversity of AICEs in Actinomycetales. Despite the large sample of genomes investigated, there is no clear evidence of AICE exchange between bacteria of the same species, of the same genus or between genera as no identical or nearly identical AICEs were detected. This observation suggests that these elements are not as promiscuous as some ICEs or conjugative plasmids found in the Firmicutes and Gram-negative bacteria, which often carry multiple antibiotic resistance genes. AICEs have never been described as vectors of such genes, preventing them from benefiting from the current anthropic selection pressure exerted by antibiotics in the environment. Alternatively, despite the 275 tested genomes, the sample size might just be too small and their respective ecological niches to diverse to allow detection of such events.
Integration and excision of the predicted AICEs seem to rely primarily (87%) on integrases of the tyrosine recombinase family. As often reported for other MGEs, predicted AICEs coding for a tyrosine recombinase often integrate into the 3′ end of a tRNA gene (73%). Tyrosine integrase AICEs for which the integration site is not a tRNA gene mostly belong to the pSLS clade, one of eight novel tyrosine integrase subfamilies. AICEs coding for a serine recombinase are rather uncommon (13%). Notably, a cluster of 8 closely related serine integrases seem to catalyze AICE integration into a distinct and unique tRNA Leu gene in several species of Mycobacterium. To date, only a few examples of MGEs, mostly bacteriophages from Mycobacteria, have been shown to use a serine integrase for integration into or near tRNA genes.14,15
Rolling circle replication (RCR) was found to be necessary for successful conjugative transfer of pSAM2, the first described AICE (RepSA replication protein).11,16 Replication is also known to occur for pMEA300 (RepAM replication protein).17 Consequently, a replication initiator gene is considered as an essential component of a canonical AICE. Indeed RepSA proteins are the most prevalent RCR proteins found in AICEs (69.4%). Phylogenetic analysis of these proteins reveals two RepSA subfamilies, RepSASAM2 and RepSAMR2. Interestingly, sequence comparison of the three conserved amino acid motifs suggests that while all RepSA proteins likely catalyze the nick formation at the double strand origin (dso) using the same mechanism, RepSASAM2 and RepSAMR2 likely exhibit significant dissimilarities for dso recognition.13,18,19 RepAM proteins are encoded by a minority (11.8%) of AICEs. Furthermore, genes encoding a Prim-pol domain-protein are often located immediately upstream of a repAM gene. Prim-pol domain proteins are likely involved in replication by acting as primases. As a consequence, the presence of Prim-pol genes can also be used to detect new classes of AICEs relying on replication proteins different from RepSA and RepAM. Out of 40 AICEs bearing a Prim-Pol gene, only 12 also bear an adjacent repAM gene, while 16 AICEs carry a SCO4618-like gene instead, which could also be involved in replication. The adjacent genes in the 12 other AICEs do not code for known RCR replication proteins and do not form a homogenous group based on sequence comparison. Nevertheless, solely based on the location of these genes, several of them could carry out replication-associated functions.
By definition, all AICEs encode a conserved FtsK-domain family transfer protein (Tra). Tra proteins are thought to be the main and sole protein required for double-stranded DNA intermycelial transfer of AICEs (TraSA) and Streptomyces conjugative plasmids (TraB). A phylogenetic analysis of AICE Tra proteins reveals that they group into six subfamilies with only one containing also a TraB protein. While TraB protein of plasmid pJVI clusters within the TraSLP1 subfamily, which contains almost exclusively AICEs, it only shares 76% identity with its closest AICE relative encoded by pSLS. Therefore, exclusively based on the comparison of their Tra proteins, emergence of new AICEs through recombination events with Streptomyces conjugative plasmids seems very unlikely.
Tra proteins could potentially promote the dissemination of unrelated genomic islands and plasmids. Acquisition of chromosomal genes, not identified as being part of any self-transmissible MGE, was shown to depend upon the presence in the donor cell of the Tra proteins encoded by Streptomyces lividans conjugative plasmid pIJ101.20 Recent work from Vogelmann et al. suggests that such events could result from the action of trans-encoded Tra proteins binding clt-like chromosomal sequences (clcs).4 Based on this observation, mobilization of genomic islands or plasmids containing clcs by an AICE or conjugative plasmid coding for a compatible Tra protein is plausible. Conceptually related mobilizable genomic islands (MGIs) were recently characterized in γ-proteobacteria.21 These MGIs rely on the recognition of their oriT by DNA processing enzymes (relaxase and auxiliary proteins forming the relaxosome) of ICEs of the SXT/R391 family for their own transfer, in addition to the ICE encoded T4CP and T4SS conjugative machinery.
T4SS-Based ICEs in Actinobacteria
While ICEs relying on a T4SS-type DNA translocation machinery for their conjugative transfer are widely distributed in Gram-negative bacteria and, to a lesser extent in Firmicutes, they appear to be scarce in actinobacteria.13,22 This discrepancy could be genuine. Conversely, it could result from the inherent and inevitable bias in the predictions introduced by using protein models based on the proteins of the widely studied prototypical conjugative plasmids found in proteobacteria.
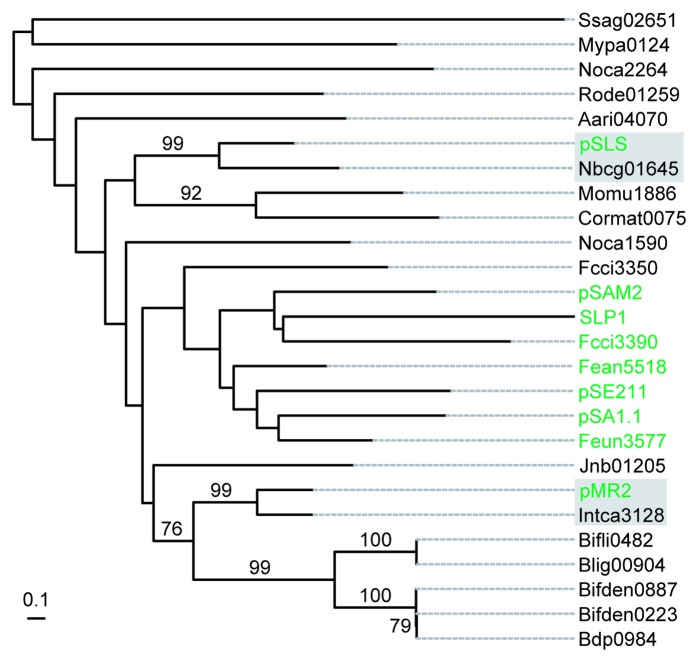
Nonetheless, actinobacteria are predicted to contain at least 17 putative T4SS-based ICEs.13 These elements were predicted in silico by seeking co-occurrences of genes coding for an integrase (Int), a TrwC-like relaxase (MOB), a VirD4-like coupling protein (T4CP) and a VirB4-like T4SS component. attL and attR attachment sites were predicted for seven out of the 17 ICEs. Most of these T4SS ICEs are site specifically integrated into the 3′ end of tRNA-encoding genes, with the exception of Nbcg01645 and Fcci3350. The excision of Fcci3350 T4SS-based ICE from Frankia sp Cci3 was confirmed by nested PCR resulting in the formation of a 71-kb circular molecule (data not shown).13 Interestingly, two actinobacterial T4SS ICEs, Intca3128 and Nbcg01645, encode tyrosine integrases that cluster with those of AICEs pMR2 and pSLS, respectively (Fig. 3). This suggests that recombination between ancestral AICEs and T4SS ICEs occurred, leading to exchange of functional modules.
Figure 3. Phylogenetic analysis of the actinobacterial T4SS-type ICE tyrosine integrases. The phylogenetic relatedness of the tyrosine integrases encoded by 17 T4SS-type ICEs (black) with those of the 9 AICEs tyrosine-integrase subfamilies (green) is represented. For simplification, only the 9 proteins encoded by the AICEs of each eponymous subfamily were used for this analysis. The relatedness of the tyrosine integrases of Intca3128 and Nbcg01645 with those of pMR2 and pSLS, respectively, is emphasized (gray shading).
Auxiliary Functions Encoded by T4SS-Based ICEs from Actinobacteria
Besides genes required for their own mobility, T4SS-based ICEs from actinobacteria also carry numerous “cargo” genes encoding a variety of putative functions identified using HMMsearch23 against all Pfam-A families from Pfam 26.0 database.24 The “cargo” genes of seven of the putative T4SS ICEs, can be classified into three groups based on their predicted functions: (1) genes coding for proteins involved in cell wall metabolism, (2) genes coding for adaptive functions and (3) genes coding for proteins involved in the translocation of a variety of molecules across the bacterial cell wall (Table S1).
The first group includes genes coding for proteins involved in the cell wall degradation such as putative CHAPS domain protein, bacteriophage peptidoglycan hydrolase and other members of the amidase family. Several genes coding for proteins involved in cell wall biosynthesis such as putative LysM domain protein, GtrA family protein and glycosyl transferase family 2 proteins are also predicted. Genes associated with adaptive functions include heavy metals resistance (putative chromate resistance protein and chromate ion transporter CHR family), type II and type III DNA restriction-modification systems, and antibiotic synthesis (aminotransferase from the DegT/DnrJ/EryC1/StrS family).25 T4SS-based ICEs from actinobacteria also carry genes involved in cell persistence and/or plasmid stabilization systems such as toxin-antitoxin and plasmid DNA partitioning loci.
To the third group appertain genes coding for secretion and transport functions such as type II secretion system and proteins of major facilitator superfamily and ATP-binding cassette (ABC) transporters that transport a wide variety of substrates such as ions, sugars, lipids, sterols, peptides, proteins and drugs across the biological membrane.
Other Conjugative MGEs and Related Elements to be Found?
Recently, T4SS-based ICEs and related elements were also more extensively sought for in all prokaryotes by Guglielmini et al.22 The predictions of this exhaustive study based on protein HMM profiles, built using proteins encoded by known conjugative plasmids, led to the identification of 335 T4SS-based ICEs in prokaryotes. Interestingly, the elements identified in actinobacteria in that study were not identified by our analyses. Conversely, none of the putative T4SS-based ICEs identified in our study was retrieved by Guglielmini and coworkers. Differences in the protein profiles used in the two studies most likely explain this observation. On one hand, to have an insight on the prevalence of T4SS-like ICEs in actinobacteria only the MOBF relaxase family was considered given that to date, the vast majority of MOB relaxases associated with conjugative plasmids in actinobacteria belong to the MOBF family and very few to the MOBQ family.13 On the other hand, all six MOB relaxase families26 were considered to identify ICEs in all prokaryotes.22 However, the 17 MOBF encoding ICEs found in actinobacteria were not identified by this latter study presumably because of the use of a more stringent protein profile for this family. Relaxases belonging to the MOBF family typically contain an N-terminal TrwC relaxase domain and a central or C-terminal DNA helicase domain. Given that the helicase domain appears to be dispensable in some instances, only the TrwC Pfam HMM protein profile responsible for the MOBF relaxases activity was used. This illustrates well some of the limitations of HMM protein profile-based studies. Protein profiles models are chosen based on assumptions of the conservation of known and previously predicted proteins. HMM protein profile-based analyses are therefore prone to the introduction of a bias, although it represents a major improvement compared with single-protein based alignment (BLAST).
Hence, despite recent extensive in silico analyses designed to identify ICEs relying on either T4SS13,22 or FtsK-like13 conjugative machineries, entire new families of more exotic types of conjugative elements could have been easily overlooked. As an example, genomes of non-actinomycete actinobacteria could host FtsK-like ICEs relying on more distant and/or unrelated proteins for their replication. In fact, an AICE-related element has been predicted in a strain of Bifidobacterium longum by using replication protein profiles not usually associated with AICEs.13 While predicted to encode a tyrosine int gene and an FtsK/SpoIIIE tra gene, this AICE-related element would replicate by means of a rep2-type replication protein.
Furthermore, most if not all ICEs characterized to date seem to be exclusively relying on a tyrosine or, more scarcely, serine recombinase, to promote their integration into and excision from a replicon. Yet, several examples of transposons, genomic islands and even viruses use DDE transposases instead to mediate similar events.27 One can wonder why such enzymes are not more frequently found associated with maintenance and mobility of ICEs. It is possible that DDE recombinases are not as well suited as tyrosine recombinases to maintain the integrity of large MGEs. It is also possible that the combination of a tyrosine recombinase and of a recombination directionality factor (Int/Xis pair) is better suited to finely tune the integration into and the excision from the chromosome of large DNA molecules.
It is tempting to speculate that many more elements related to ICEs of both superfamilies are yet unidentified. As an example, further examination of Frankia genomes reveals putative elements lacking one of the AICE canonical components. These elements could be AICEs remnants subject to genetic decay or more interestingly, be genuine MGEs, which could depend on interactions with proteins encoded by other self-transmissible mobile elements.
Predictions of ICEs and related elements in actinobacteria have enabled to gain better insights on the distribution, diversity and evolution of these MGE. Interestingly, ICEs encoding two mechanistically different DNA translocation machineries are present in actinobacteria. However, despite the apparent simplicity of their “conjugative apparatus,” AICEs seem to have the biggest share of the gene trade in actinomycetes.
Supplementary Material
Acknowledgments
We are grateful to Nicolas Carraro for helpful comments on the manuscript. V.B. holds a Canada Research Chair in Bacterial Molecular Genetics. E.B. is the recipient of an Alexander Graham Bell Canada Graduate Scholarship from NSERC.
Glossary
Abbreviations:
- ICE
integrating conjugative element
- AICE
actinomycete integrating conjugative element
- T4SS
type IV secretion system
- T4CP
type IV coupling protein
- MGE
mobile genetic element
- HMM
Hidden Markov model
- clt
cis-acting locus of transfer
- clcs
clt-like chromosomal sequences
Footnotes
Previously published online: www.landesbioscience.com/journals/mge/article/20498
References
- 1.Burrus V, Pavlovic G, Decaris B, Guédon G. Conjugative transposons: the tip of the iceberg. Mol Microbiol. 2002;46:601–10. doi: 10.1046/j.1365-2958.2002.03191.x. [DOI] [PubMed] [Google Scholar]
- 2.Wozniak RA, Waldor MK. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol. 2010;8:552–63. doi: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]
- 3.de la Cruz F, Frost LS, Meyer RJ, Zechner EL. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol Rev. 2010;34:18–40. doi: 10.1111/j.1574-6976.2009.00195.x. [DOI] [PubMed] [Google Scholar]
- 4.Vogelmann J, Ammelburg M, Finger C, Guezguez J, Linke D, Flötenmeyer M, et al. Conjugal plasmid transfer in Streptomyces resembles bacterial chromosome segregation by FtsK/SpoIIIE. EMBO J. 2011;30:2246–54. doi: 10.1038/emboj.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alvarez-Martinez CE, Christie PJ. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev. 2009;73:775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fronzes R, Christie PJ, Waksman G. The structural biology of type IV secretion systems. Nat Rev Microbiol. 2009;7:703–14. doi: 10.1038/nrmicro2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smillie C, Garcillán-Barcia MP, Francia MV, Rocha EP, de la Cruz F. Mobility of plasmids. Microbiol Mol Biol Rev. 2010;74:434–52. doi: 10.1128/MMBR.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biller SJ, Burkholder WF. The Bacillus subtilis SftA (YtpS) and SpoIIIE DNA translocases play distinct roles in growing cells to ensure faithful chromosome partitioning. Mol Microbiol. 2009;74:790–809. doi: 10.1111/j.1365-2958.2009.06893.x. [DOI] [PubMed] [Google Scholar]
- 9.Kaimer C, González-Pastor JE, Graumann PL. SpoIIIE and a novel type of DNA translocase, SftA, couple chromosome segregation with cell division in Bacillus subtilis. Mol Microbiol. 2009;74:810–25. doi: 10.1111/j.1365-2958.2009.06894.x. [DOI] [PubMed] [Google Scholar]
- 10.Begg KJ, Dewar SJ, Donachie WD. A new Escherichia coli cell division gene, ftsK. J Bacteriol. 1995;177:6211–22. doi: 10.1128/jb.177.21.6211-6222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hagège J, Pernodet JL, Friedmann A, Guérineau M. Mode and origin of replication of pSAM2, a conjugative integrating element of Streptomyces ambofaciens. Mol Microbiol. 1993;10:799–812. doi: 10.1111/j.1365-2958.1993.tb00950.x. [DOI] [PubMed] [Google Scholar]
- 12.te Poele EM, Bolhuis H, Dijkhuizen L. Actinomycete integrative and conjugative elements. Antonie Van Leeuwenhoek. 2008;94:127–43. doi: 10.1007/s10482-008-9255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghinet MG, Bordeleau E, Beaudin J, Brzezinski R, Roy S, Burrus V. Uncovering the prevalence and diversity of integrating conjugative elements in actinobacteria. PLoS One. 2011;6:e27846. doi: 10.1371/journal.pone.0027846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Askora A, Kawasaki T, Fujie M, Yamada T. Resolvase-like serine recombinase mediates integration/excision in the bacteriophage φRSM. J Biosci Bioeng. 2011;111:109–16. doi: 10.1016/j.jbiosc.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 15.Pope WH, Jacobs-Sera D, Russell DA, Peebles CL, Al-Atrache Z, Alcoser TA, et al. Expanding the diversity of mycobacteriophages: insights into genome architecture and evolution. PLoS One. 2011;6:e16329. doi: 10.1371/journal.pone.0016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagège J, Boccard F, Smokvina T, Pernodet JL, Friedmann A, Guérineau M. Identification of a gene encoding the replication initiator protein of the Streptomyces integrating element, pSAM2. Plasmid. 1994;31:166–83. doi: 10.1006/plas.1994.1018. [DOI] [PubMed] [Google Scholar]
- 17.te Poele EM, Kloosterman H, Hessels GI, Bolhuis H, Dijkhuizen L. RepAM of the Amycolatopsis methanolica integrative element pMEA300 belongs to a novel class of replication initiator proteins. Microbiology. 2006;152:2943–50. doi: 10.1099/mic.0.28746-0. [DOI] [PubMed] [Google Scholar]
- 18.Orozco BM, Gladfelter HJ, Settlage SB, Eagle PA, Gentry RN, Hanley-Bowdoin L. Multiple cis elements contribute to geminivirus origin function. Virology. 1998;242:346–56. doi: 10.1006/viro.1997.9013. [DOI] [PubMed] [Google Scholar]
- 19.Orozco BM, Hanley-Bowdoin L. Conserved sequence and structural motifs contribute to the DNA binding and cleavage activities of a geminivirus replication protein. J Biol Chem. 1998;273:24448–56. doi: 10.1074/jbc.273.38.24448. [DOI] [PubMed] [Google Scholar]
- 20.Pettis GS, Cohen SN. Transfer of the plJ101 plasmid in Streptomyces lividans requires a cis-acting function dispensable for chromosomal gene transfer. Mol Microbiol. 1994;13:955–64. doi: 10.1111/j.1365-2958.1994.tb00487.x. [DOI] [PubMed] [Google Scholar]
- 21.Daccord A, Ceccarelli D, Burrus V. Integrating conjugative elements of the SXT/R391 family trigger the excision and drive the mobilization of a new class of Vibrio genomic islands. Mol Microbiol. 2010;78:576–88. doi: 10.1111/j.1365-2958.2010.07364.x. [DOI] [PubMed] [Google Scholar]
- 22.Guglielmini J, Quintais L, Garcillán-Barcia MP, de la Cruz F, Rocha EP. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet. 2011;7:e1002222. doi: 10.1371/journal.pgen.1002222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23:205–11. doi: 10.1142/9781848165632_0019. [DOI] [PubMed] [Google Scholar]
- 24.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38(Database issue):D211–22. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahlert J, Distler J, Mansouri K, Piepersberg W. Identification of stsC, the gene encoding the L-glutamine:scyllo-inosose aminotransferase from streptomycin-producing Streptomycetes. Arch Microbiol. 1997;168:102–13. doi: 10.1007/s002030050475. [DOI] [PubMed] [Google Scholar]
- 26.Garcillán-Barcia MP, Francia MV, de la Cruz F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol Rev. 2009;33:657–87. doi: 10.1111/j.1574-6976.2009.00168.x. [DOI] [PubMed] [Google Scholar]
- 27.Nesmelova IV, Hackett PB. DDE transposases: Structural similarity and diversity. Adv Drug Deliv Rev. 2010;62:1187–95. doi: 10.1016/j.addr.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.