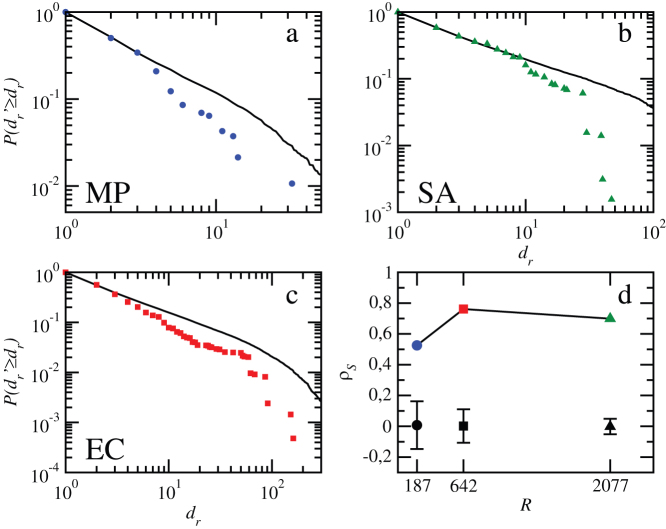
Figure 1. Damage in cascades triggered by individual reactions.
(a–c). Cumulative probability distribution functions of damages in M. pneumoniae, E. coli, and S. aureus. Results are compared with damages produced in degree-preserving randomized versions of the metabolic networks in order to discount structural effects. In each case, the solid black curve is the average over 100 realizations. Results for S. aureus and an older version of E. coli were already presented in Ref.(19). The results of the Kolmogorov-Smirnov tests are given in terms of the K-S statistic and its associated significance level: 0.095/0.07, 0.086/0.0002, and 0.079/1.4 · 10−11 for M. pneumoniae, S. aureus, and E. coli respectively. With a significance value of α = 0.05, distributions of damages can be considered not consistent with those for randomized variants, except for M. pneumoniae. (d) Spearman's rank correlation coefficient ρS between predictors and damages, plotted against metabolic network size (number of reactions R). Results are compared to random reshuffling of the predictor value associated to reactions (100 realizations for each organism). Average Spearman's rank correlation coefficients for the randomizations appear in black, and error bars delimit the maximum and the minimum values obtained.