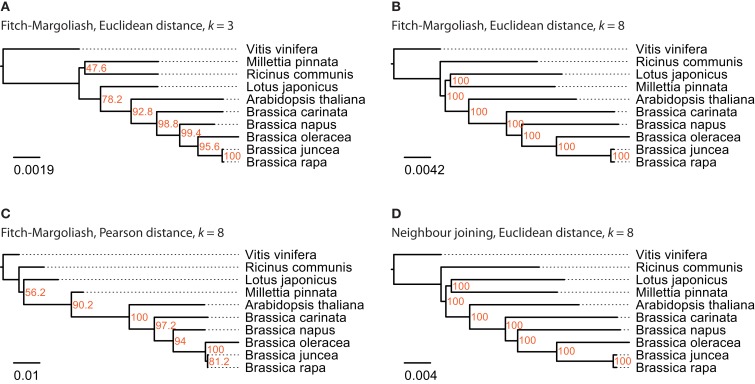
Figure 5.
The phylogenetic trees were generated applying different methods on FCGRs of available mitochondrial genome sequence data of the Rosids taxon. In (A–C) the Fitch–Margoliash algorithm was used to calculate trees for 500 re-sampled datasets. Subsequently, a consensus tree was built and branch lengths were calculated based on the fixed consensus tree. The method used for the distance calculation and the resolution of the FCGRs are given on top of the trees. In (D) the Neighbor joining algorithm was used to calculate the tree.