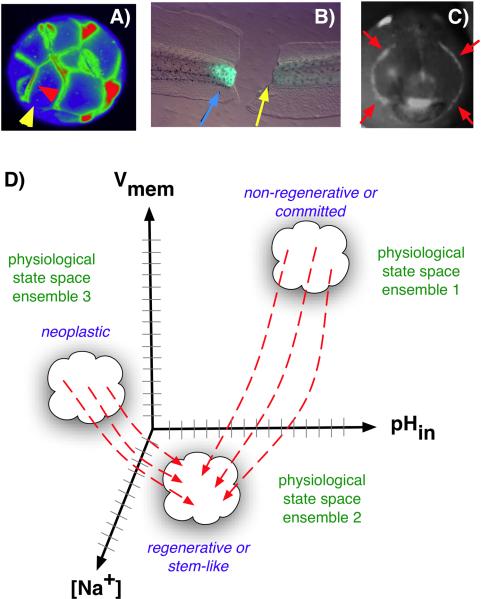
Figure 2.
Voltage gradients in vivo. A: Fluorescent voltage reporter dyes allow characterization of physiological gradients in vivo, such as this image of a 16-cell frog embryo that simultaneously reveals cells' potential levels (blue = hyperpolarized, red = depolarized) in vivo, as well as domains of distinct Vmem around a single blastomere's surface (compare the side indicated by the yellow arrowhead with the one indicated by the red arrowhead). B: Gradients of transmembrane potential demarcate important tissue domains, such as the depolarized region shortly after tail amputation in Xenopus laevis tadpoles (blue arrowhead), which will give rise to the regeneration bud, and can reveal non-regenerative conditions when the appropriate physiological state had not been achieved, or was experimentally blocked (yellow arrowhead). C: Isopotential cell fields can also demarcate subtle prepatterns existing in tissues, such as the hyperpolarized domains (red arrowheads) that presage the expression of regulatory genes such as Frizzled during frog embryo craniofacial development [13]. It is necessary to gain a quantitative understanding of the bioelectric code – to map out the linkage between physiological state with cell behavior outcomes, as a prelude to a full understanding of how 3-dimensional patterning information is stored in physiological properties of tissue. D: One hypothesis is that cell types (e.g. proliferative, or neoplastic, or undifferentiated) cluster in a multi-dimensional state space in which each axis defines the value of a physiological parameter. Additional axes (not shown) could include levels of other ions (chloride, potassium), nuclear membrane potential, cell surface charge (zeta potentials), etc. Once appropriate data are gathered, cells could be moved from their current state to a desired state by pharmacological and molecular-genetic changes shifting them along each axis, toward a different ensemble within the state space as needed for a given biomedical application.