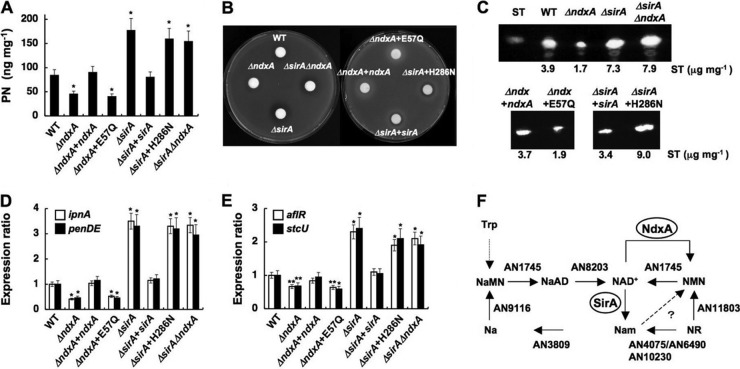
Fig 6.
NdxA requirement for maximal secondary metabolite production. (A) Penicillin G (PN) production after 72 h in culture. (B) Bacterial growth inhibition assay shows penicillin production by A. nidulans. (C) Sterigmatocystin production after 72 h in culture. (D and E) Transcripts of secondary metabolite genes in cells quantified by PCR after 72 h in culture. WT, wild type; ΔndxA, ΔNdxA; ΔndxA+ndxA, ΔNdxA2 strain transformed with pNdxA; ΔndxA+E57Q, ΔNdxA2 strain transformed with pNdxA-E57Q; ΔsirA, ΔSirA; ΔsirA+sirA, ΔSirA2 strain transformed with pSirA; ΔsirA+H286N, ΔSirA2 strain transformed with pSirA-H286N; ΔsirAΔndxA, ΔSirA ΔNdxA. Data are means ± standard deviations (n = 3). *, P < 0.005 versus WT. (F) NAD+ synthesis in A. nidulans. Predicted genes for nicotinamide riboside (NR) salvage pathways for NAD+ synthesis and for de novo synthetic pathway of NAD+ are presented as gene identities. NaMN, nicotinic acid mononucleotide; NaAD, nicotinic acid adenine dinucleotide; NMN, nicotinamide mononucleotide; Na, nicotinic acid; Nam, nicotinamide. NdxA counteracts NMN adenylyltransferase (AN1745) in that they catalyze NMN+ and NAD+ interconversion.