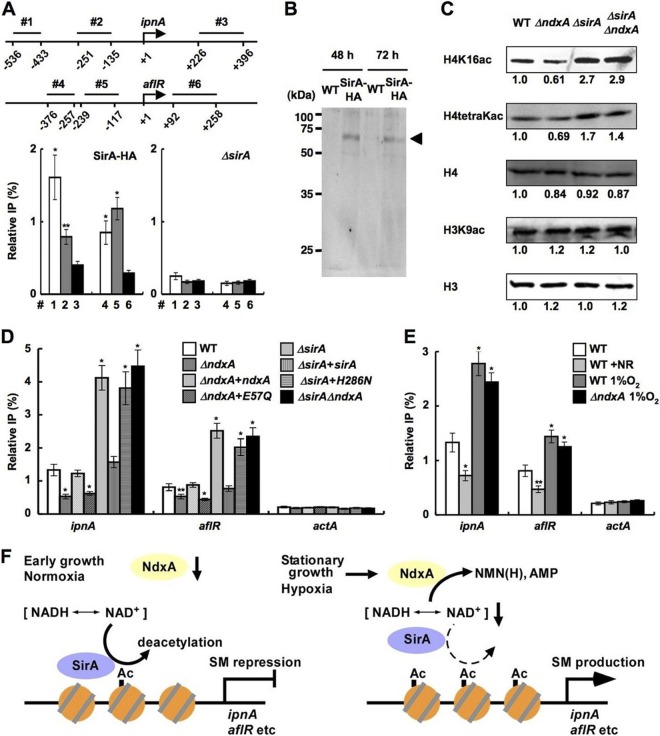
Fig 9.
NdxA represses histone deacetylation. (A) ChIP analyses indicate SirA association with ipnA and aflR gene promoters in A. nidulans cells cultured for 48 h. Top panel, positions (1 to 6) of immunoprecipitated DNA. Data are means ± standard deviations (n = 3). *, P < 0.005, and **, P < 0.01, versus ΔsirA. (B) Western blot analysis of cell extracts of the strain producing SirA-HA. Anti-HA antibody detected levels of SirA-HA similar between early stationary (48-h) and stationary (72-h) growth phases. Arrowhead, SirA-HA. (C) Western blotting detected acetylated histones in nuclear fractions of cells cultured for 72 h. H3 and H4, total histones H3 and H4. Representative results of four repeated experiments are presented with relative signal intensity under bands. (D and E) ChIP findings of effects of ndxA and sirA (D) and of nicotinamide riboside and hypoxia (E) on H4K16 acetylation at ipnA, aflR, and actA gene promoters in strains cultured for 72 h. Relative IP represents signal ratio between precipitated and input DNA. WT, wild type; ΔndxA, ΔNdxA; ΔndxA+ndxA, ΔNdxA2 strain transformed with pNdxA; ΔndxA+E57Q, ΔNdxA2 strain transformed with pNdxA-E57Q; ΔsirA, ΔSirA; ΔsirA+sirA, ΔSirA2 strain transformed with pSirA; ΔsirA+H286N, ΔSirA2 strain transformed with pSirA-H286N; ΔsirAΔndxA, ΔSirA ΔNdxA. Data are means ± standard deviations (n = 3). *, P < 0.005, and **, P < 0.01, versus WT. (F) Model of negative epigenetic control by NdxA through NAD(H) hydrolysis.