Abstract
The bacterial pathogen Pseudomonas syringae pv. tomato DC3000 must detoxify plant-produced hydrogen peroxide (H2O2) in order to survive in its host plant. Candidate enzymes for this detoxification include the monofunctional catalases KatB and KatE and the bifunctional catalase-peroxidase KatG of DC3000. This study shows that KatG is the major housekeeping catalase of DC3000 and provides protection against menadione-generated endogenous H2O2. In contrast, KatB rapidly and substantially accumulates in response to exogenous H2O2. Furthermore, KatB and KatG have nonredundant roles in detoxifying exogenous H2O2 and are required for full virulence of DC3000 in Arabidopsis thaliana. Therefore, the nonredundant ability of KatB and KatG to detoxify plant-produced H2O2 is essential for the bacteria to survive in plants. Indeed, a DC3000 catalase triple mutant is severely compromised in its ability to grow in planta, and its growth can be partially rescued by the expression of katB, katE, or katG. Interestingly, our data demonstrate that although KatB and KatG are the major catalases involved in the virulence of DC3000, KatE can also provide some protection in planta. Thus, our results indicate that these catalases are virulence factors for DC3000 and are collectively required for pathogenesis.
INTRODUCTION
Pseudomonas syringae pv. tomato DC3000 is a hemibiotrophic Gram-negative bacterium that is the causative agent of bacterial speck disease in susceptible cultivars of tomato (Solanum lycopersicum) and a virulent pathogen of the model plant Arabidopsis thaliana. P. syringae can live both as an epiphyte and in the aerial tissues of its host plants. It infects its host through wounds or natural openings such as stomata and lives extracellularly in the apoplast. Plants can recognize conserved molecules on invading microbes called pathogen (microbe)-associated molecular patterns (PAMPs/MAMPs) using pattern recognition receptors (PRRs). This recognition leads to activation of a set of host immune responses aimed at the removal or restriction of the invading microbe. These responses are collectively termed PAMP-triggered immunity (PTI) (2, 40). In order to be a successful pathogen, P. syringae must suppress, tolerate, or evade the PTI response of its host plant.
One of the earliest outputs of PTI in Arabidopsis is an oxidative burst that occurs within minutes of pathogen detection (30, 40). This oxidative burst consists of a rapid and transient production of reactive oxygen species (ROS) such as hydrogen peroxide (H2O2) and superoxide (O2−). In Arabidopsis, the apoplastic oxidative burst appears to be, at least in part, due to activation of the plasma membrane-localized NADPH oxidases AtrbohD and AtrbohF (6, 29, 50) and apoplastic and peroxisomal peroxidases (10, 36). In addition to the apoplastic oxidative burst, plant ROS are also produced by perturbations in metabolic processes, particularly the uncoupling or inhibition of photosynthesis and photorespiration (23).
ROS play multiple roles in plant defense, as they can be signaling molecules, form oxidative cross-linkages in the plant cell wall to prevent pathogen ingress, or impact bacterial growth (46). The high levels of ROS that accumulate in the apoplast during the oxidative burst could potentially kill invading microbes directly. ROS are toxic for bacteria, as they cause DNA breakage, oxidation of critical enzymes, and lipid peroxidation leading to defective DNA replication and malfunction of proteins and membranes (5, 18, 25). Consequently, ROS, including H2O2, could limit the ability of DC3000 to multiply and survive in the plant apoplast. It is therefore critical for P. syringae to suppress and/or detoxify these ROS in order to be pathogenic in its host plant.
Like other pathogens, P. syringae uses multiple strategies to overcome plant innate immunity and to cause disease in host plants. A major virulence mechanism of DC3000 is the injection of a battery of virulence proteins called type III effectors into the plant cell by using the type III secretion system. Many of these type III effectors suppress plant innate immunity, and several have been shown to interact with and inhibit PRRs and other PTI signaling components (1). This inhibition attenuates but does not eliminate the production of the oxidative burst; therefore, P. syringae must retain the ability to detoxify apoplastic H2O2. Several bacterial pathogens have been shown to use catalases to detoxify H2O2 by converting it to water and oxygen. For example, a catalase of the human pathogen Pseudomonas aeruginosa is induced in response to oxidative stress and provides resistance to H2O2 (3, 34). In addition, the catalases of phytopathogenic or plant-associated bacteria such as P. syringae pv. phaseolicola (37), Pseudomonas putida (24), and Sinorhizobium meliloti (19) are also induced in planta, indicating that bacterial catalases may be involved in the detoxification of plant-produced ROS.
Most pathogenic P. syringae strains, including the sequenced strains P. syringae pv. phaseolicola 1448a and P. syringae pv. syringae B728a, have 5 catalases. These are the heme-containing catalases katA, katB, katE, and katG and a nonheme catalase (12, 22). However, DC3000 contains only three of these catalase genes: katB (PSPTO_3582), katE (PSPTO_5263), and katG (PSPTO_4530) (4). KatB and KatE are monofunctional catalases, whose substrate is exclusively H2O2, while KatG is a bifunctional catalase, also known as hydroperoxidase, which exhibits both catalase and peroxidase activity (42). Bifunctional catalases are able to use organic peroxides as substrates as well as H2O2. Different species of phytopathogenic bacteria have been shown to use different catalases to assist in their virulence in planta. For instance, Agrobacterium tumefaciens uses KatA, Xanthomonas campestris pv. campestris uses KatA and KatG, and Xanthomonas axonopodis pv. citri uses KatE (21, 45, 49). Interestingly, the symbiotic bacterium Sinorhizobium meliloti uses both mono- and bifunctional catalases to aid its symbiosis (20, 41). It is not known which, if any, of DC3000's catalases protect it from plant-derived ROS or if they play an important role in its virulence. Here, we assess the relative roles of the catalases of DC3000 and show that catalases are important P. syringae virulence factors.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All Escherichia coli strains were grown in LM medium with appropriate antibiotics at 37°C. Pseudomonas syringae pv. tomato DC3000 strains were grown in King's B (KB) medium (26) or Casamino Acids M9 medium at 30°C. Antibiotics were used at the following concentrations: ampicillin (Ap), 100 μg ml−1; gentamicin (Gm), 1 μg ml−1; kanamycin (Km), 50 μg ml−1; rifampin (Rif), 100 μg ml−1; spectinomycin (Sp), 50 μg ml−1; and tetracycline (Tc), 20 μg ml−1.
DNA manipulation.
Well-described protocols were followed for standard DNA manipulations (39). Restriction enzymes, T4 ligase, and DNA polymerase were purchased from New England BioLabs (Beverly, MA). The thermostable Pfu DNA polymerase (Stratagene, La Jolla, CA) was used to amplify the desired DNA fragments by PCR. Oligonucleotide primers were ordered from Integrated DNA Technologies (Coralville, IA). For cloning using Gateway technology, the PCR-amplified DNA regions were cloned into the pENTR/D-TOPO vector (Invitrogen, Carlsbad, CA). The resulting pENTR constructs were recombined with Gateway destination vectors by LR reaction using LR Clonase (Invitrogen) according to the manufacturer's instructions.
Construction of DC3000 catalase mutants.
An unmarked mutagenesis strategy modified from House et al. (17) was used to construct DC3000 catalase mutants. Briefly, upstream or downstream DNA regions of each catalase gene were PCR amplified with Pfu polymerase (Stratagene) and then cloned into the pENTR/D-TOPO vector (Invitrogen, Carlsbad, CA). The upstream and downstream DNA regions of katB were amplified with primer pairs P2587 (5′-CACCAAGGCTTACCGGGGCTGAATG-3′)/P2588 (5′-CGGCATTTTGAATCCCTCATATTT-3′) and P2593 (5′-CACCAAGGTCAAGTCACTGGCTG-3′)/P2592 (5′-AATGGCACCAGTGCGTTCATC-3′), respectively; those of katE with primer pairs P2594 (5′-CACCAGCGCGATGGCAGTGACG-3′)/P2595 (5′-AGCCATCAGAAACTCCTTATC-3′) and P2596 (5′-CACCGCTTGATCTAAACGCCAG-3′)/P2597 (5′-ATGGCTGATTTTCTACTG-3′), respectively; and those of katG with primer pairs P2598 (5′-CACCCCGGAAGATTCTCGCC-3′)/P2599 (5′-TGACATGCTTGATACACC-3′) and P2600 (5′-CACCTGAACCTGGACCGTTTCG-3′)/P2601 (5′-ATGCTCCATTTATTCCGC-3′), respectively. The resulting pENTR constructs were recombined with suicidal destination vector pMK2016 or pMK2017, respectively, using LR Clonase according to the manufacturer's instructions. The pMK2016 and pMK2017 derivatives were recombined into the DC3000 chromosome by triparental or biparental mating. The conjugating plasmid pBH474, which constitutively expresses flp recombinase, was then introduced into the resulting strains to excise the targeted catalase genes. The strains were cured of the plasmid, and colonies sensitive to both tetracycline and spectinomycin were identified as catalase deletion mutants. The flp recombinase-dependent deletion results in a 230-bp scar between the flanking sequences of the deleted gene (17). All mutants were confirmed by sequencing the PCR-amplified scar using primers to the flanking sequences of the genes.
For the complementation strains, catalases under the control of their native promoters were expressed individually using the Tn7 expression system (7). The catalase genes with their native promoters were amplified using P3391 (5′-CACCACTAGTACGACACTGTCATCTTCCTG-3′) and P2621 (5′-GTATAGCCTTGTTCGTC-3′) for katB; P2978 (5′-CACCACTAGTTCAGCGCAGTGGCAGGACGC-3′) and P3393 (5′-AGTAAGCTTCTCCTTATCTCGACCCAATC-3′) for katE; and P2980 (5′-CACCACTAGTTGGCAGAGCCCTTCGCTACGC-3′) and P2826 (5′-GGTGGATGTGCATTTGAAG-3′) for katG. The PCR fragments were cloned into pENTR, resulting in pLN4540 (katB), pLN4543 (katE), and pLN4539 (katG). The above plasmids were recombined with the Gateway compatible Tn7 vector pUC18T mini-Tn7T-Gm-GW, resulting in Tn7 constructs pLN4225 (katB), pLN4223 (katE), and pLN4224 (katG). The resulting Tn7 constructs were coelectroporated with the helper plasmid pTNS2 into their respective mutant strains. The gentamicin-resistant colonies were selected for Tn7 transposition events. Plasmids and strains used in this study are listed in Table 1.
Table 1.
Plasmids and strains used in this study
| Strain or plasmid | Characteristics | Reference or source |
|---|---|---|
| Strains | ||
| E. coli DB3.1 | F−gyrA462 endA1Δ(sr1-recA)mcrB mrr hsdS20(rB− mB−)supE44 ara-14 galK2 lacY1 proA2 rpsL20(Smr)xyl-5λ−leu mtl-1 | Invitrogen |
| E. coli DH5α | supE44ΔlacU169(ϕ80lacZΔM15)hsdR17 recA1 endA1 gyrA96 thi-1 relA1, Nalr | 14; Life Technologies |
| E. coli S17-1 λpir | Tpr Smr recA thi pro hsdr− m+ RP4::2-Tc::Mu::Km Tn7λ[ι] pir | K. N. Timmis |
| P. syringae pv. tomato DC3000 | Wild type; spontaneous Rifr | 9 |
| UNL204 | DC3000 unmarked mutant lacking katB, Rifr | This work |
| UNL211 | DC3000 unmarked mutant lacking katB and katE, Rifr | This work |
| UNL213 | DC3000 unmarked mutant lacking katE, Rifr | This work |
| UNL222 | DC3000 unmarked mutant lacking katG, Rifr | This work |
| UNL223 | DC3000 unmarked mutant lacking katB and katG, Rifr | This work |
| UNL225 | DC3000 unmarked mutant lacking katE and katG, Rifr | This work |
| UNL254 | DC3000 unmarked mutant lacking katB, katE, and katG; Rifr | This work |
| Plasmids | ||
| pBH474 | Broad-host-range plasmid harboring sacB and constitutively expressing flp, sucrose sensitive, Gmr | 17 |
| pENTR/D-TOPO | Gateway system donor vector, Kmr | Invitrogen |
| pGEX5x-1 | Vector expressing GST fusion proteins, Apr | GE Healthcare Life Sciences |
| pLN3083 | pENTR derivative carrying downstream of katB, Kmr | This work |
| pLN3084 | pENTR derivative carrying upstream of katB, Kmr | This work |
| pLN3085 | pENTR derivative carrying downstream of katE, Kmr | This work |
| pLN3086 | pENTR derivative carrying upstream of katE, Kmr | This work |
| pLN3087 | pENTR derivative carrying downstream of katG, Kmr | This work |
| pLN3088 | pENTR derivative carrying upstream of katG, Kmr | This work |
| pLN3095 | pMK2016 derivative carrying downstream of katB, Spr | This work |
| pLN3096 | pMK2017 derivative carrying upstream of katB, Tcr | This work |
| pLN3097 | pMK2017 derivative carrying downstream of katE, Tcr | This work |
| pLN3098 | pMK2016 derivative carrying upstream of katB, Spr | This work |
| pLN3099 | pMK2017 derivative carrying downstream of katG, Tcr | This work |
| pLN3100 | pMK2016 derivative carrying upstream of katG, Spr | This work |
| pLN3357 | pENTR derivative carrying katE-HA with its native promoter, Kmr | This work |
| pLN3358 | pENTR derivative carrying katG-HA with its native promoter, Kmr | This work |
| pLN3364 | pENTR derivative carrying katB-HA with its native promoter, Kmr | This work |
| pLN3365 | pUC18T mini-Tn7T-Gm-GW derivative carrying katB-HA with its native promoter, Gmr | This work |
| pLN4180 | pGEX5x-1 derivative carrying katB ORF fused with GST at N terminus, Apr | This work |
| pLN4181 | pGEX5x-1 derivative carrying katE ORF fused with GST at N terminus, Apr | This work |
| pLN4182 | pGEX5x-1 derivative carrying katG ORF fused with GST at N terminus, Apr | This work |
| pLN4379 | pUC18T mini-Tn7T-Gm-GW derivative carrying katE-HA with its native promoter, Apr Gmr | This work |
| pLN4380 | pUC18T mini-Tn7T-Gm-GW derivative carrying katG-HA with its native promoter, Apr Gmr | This work |
| pLN4381 | pUC18T mini-Tn7T-Gm-GW derivative carrying katB-HA with its native promoter, Apr Gmr | This work |
| pLN4539 | pENTR derivative carrying katG with its native promoter, Kmr | This work |
| pLN4540 | pENTR derivative carrying katB with its native promoter, Kmr | This work |
| pLN4541 | pUC18T mini-Tn7T-Gm-GW derivative carrying katG with its native promoter, Apr Gmr | This work |
| pLN4542 | pUC18T mini-Tn7T-Gm-GW derivative carrying katB with its native promoter, Apr Gmr | This work |
| pLN4543 | pENTR derivative carrying katE with its native promoter, Kmr | This work |
| pLN4544 | pUC18T mini-Tn7T-Gm-GW derivative carrying katE with its native promoter, Apr Gmr | This work |
| pMK2016 | oriV oriTColE1 with FRTa cassette, Spr | 17 |
| pMK2017 | oriVR6K oriTRP4 with FRT cassette, Tcr | 17 |
| pRK2013 | Helper plasmid, Kmr | 38 |
| pTNS2 | Tn7 helper plasmid, Apr | 7 |
| pUC18T mini-Tn7T-Gm-GW | Tn7 destination vector compatible with Gateway cloning, Apr Gmr | 7 |
FRT, flippase recognition target.
Hydrogen peroxide inhibition zone assay.
The H2O2 growth inhibition assays were performed as described by Xu and Pan (49), with modifications. Briefly, DC3000 strains were grown overnight at 30°C on KB plates. The cultures were resuspended in liquid KB medium, adjusted to an optical density at 600 nm (OD600) of 0.3, and mixed with 0.4% (wt/vol) agar at a ratio of 1:10 (vol/vol). Five milliliters of the mixture was layered on plates containing 20 ml of KB agar. A filter disk of 0.5 cm in diameter was then placed in the center of the plate, and 10 μl of 3% H2O2 (vol/vol) was applied to the disk. The plate was incubated at 30°C overnight. Pictures were taken after incubation, and the diameters of the inhibition zones were measured.
Menadione growth inhibition assay.
DC3000 strains grown overnight on KB plates were resuspended in liquid KB medium and adjusted to an OD600 of 0.1. Tenfold serial dilutions were made in liquid KB medium, and 5 μl of each dilution was spotted onto KB plates with or without 0.2 mM menadione. Pictures were taken after incubation at 30°C for 2 days.
In vitro catalase assay.
Crude total protein extracts of DC3000 strains were used to determine total catalase activity. DC3000, the catalase mutants, and the catalase mutants expressing catalases under the control of their native promoters in the Tn7 system were grown in 5 ml of liquid M9 Casamino Acids medium at 30°C to stationary phase. Cells were harvested from 2 ml of culture, and total protein was extracted by sonication. Protein concentration was determined using a Bradford assay, and catalase activity was determined using the catalase fluorometric detection kit (Assay Designs) according to the manufacturer's instructions.
Catalase assay for purified recombinant catalases.
All three DC3000 catalase genes were cloned into pGEX-5X-1 as N-terminal glutathione S-transferase (GST) fusions. katB was amplified with primers P3742 (5′-CACCGGATCCCCATGCCGTTATTAAACTGGTC-3′) and P3743 (5′-AGTACTCGAGTCAGTCTTTCAGGCTGGCAG-3′); katE with primers P3744 (5′-CACCGGATCCCCATGGCTAGCAAGAAAGCACC-3′) and P3745 (5′-AGTAGTCGACTCAAGCCGGAACAGCTTTGG-3′); and katG with primers P3746 (5′-CACCGGATCCCCATGTCAACTGAATCGAAATG-3′) and P3747 (5′-AGTACTCGAGTCAAGCGAGGTCGAAACGGT-3′). The above DNA fragments were cloned into the BamHI and XhoI sites of pGEX-5X-1, resulting in pLN4180, pLN4181, and pLN4182, respectively. The GST-tagged catalases were induced in E. coli BL21 cells with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Cells were lysed by sonication in 1× phosphate-buffered saline (PBS) with Complete EDTA-free protease inhibitor (Roche), and 0.5% (vol/vol) Triton X-100 was added. GST-tagged proteins were purified using the BugBuster GST-bind purification kit (Novagen) according to the manufacturer's instructions. Catalase-specific activity was determined for the purified proteins in 60 mM KH2PO4 (pH 7.5) with 8.8 mM H2O2. One unit of activity is defined as the amount of enzyme that will decompose 1.0 μmol of H2O2 per min at pH 7.5, as measured by the rate of decrease of A240.
Analysis of catalase gene expression.
Catalase mRNA levels were measured by semiquantitative reverse transcription (RT)-PCR. For transcription analysis in culture, DC3000 was grown in liquid M9 Casamino Acids medium at 30°C to an OD600 of 0.3 and H2O2 was added to a final concentration of 1 mM. Samples were taken at the indicated time points and cells harvested and flash frozen in liquid nitrogen. For expression of DC3000 catalase genes in planta, 2 × 108 cells ml−1 of DC3000 in 10 mM MgCl2 was syringe infiltrated into Arabidopsis Col-0 leaves. Leaf disks of 1 cm2 were harvested at the indicated times and flash frozen in liquid nitrogen. RNA was extracted using the RNeasy plant mini spin kit (Qiagen) with on-column DNase digestion according to the manufacturer's instructions. RNA was quantified and cDNA synthesized using Retroscript (AMBION) with random decamer priming. PCR was performed using an icycler (Bio-Rad) with IQ-SYBR-green supermix (Bio-Rad). katB was amplified with primers P4008 (5′-CGACCTGAGCAAGGCGACCG-3′) and P4009 (5′-GCGCAGGGTTTCTGGCGAGT-3′); katE with primers P4010 (5′-CAGTGGACGGCGGCTTCGAG-3′) and P4011 (5′-GCGCTCGACCTTGCCCAACT-3′); and katG with primers P4012 (5′-ACGTCGGGCAGAGCAAGCAC-3′) and P4013 (5′-TCGTTGGCACCCGAAGTCGC-3′). RNA levels were normalized with 16S rRNA using primers P4006 (5′-GATCCAGCCATGCCGCGTGT-3′) and P4007 (5′-GCGGCTGCTGGCACAGAGTT-3′). Relative RNA levels were calculated using the formula 10(ΔCT/3) × 100, where ΔCT is the difference in cycle threshold between the catalase and 16S rRNA. Fold increase of expression compared to that at time zero was then determined.
Analysis of catalase protein expression.
The Tn7 expression system was used to express catalases tagged with a hemagglutinin (HA) epitope at their C termini under the control of their native promoters in the catalase single mutants (7). Catalase genes fused to the HA epitope with their native promoters were cloned into pENTR/D-TOPO, resulting in plasmids pLN3359 (katB-HA), pLN3357 (katE-HA), and pLN3358 (katG-HA). katB-HA was amplified with primers P2976 (5′-CACCACTAGTCATCAAATAGCCACTAACCG-3′) and P2977 (5′-GATCAAGCTTTCAAGCGTAATCTGGAACATCGTATGGGTAGTCTTTCAGGCTGGCAGCCAG-3′); katE-HA with primers P2978 (5′-CACCACTAGTTCAGCGCAGTGGCAGGACGC-3′) and P2979 (5′-ATCAAGCTTTCAAGCGTAATCTGGAACATCGTATGGGTAAGCCGGAACAGCTTTGGCCTTC-3′); and katG-HA with primers P2980 (5′-CACCACTAGTTGGCAGAGCCCTTCGCTACGC-3′) and P2981 (5′-GATCAAGCTTTCAAGCGTAATCTGGAACATCGTATGGGTAAGCGAGGTCGAAACGGTCCAG-3′). pLN3359 (katB-HA), pLN3357 (katE-HA), and pLN3358 (katG-HA) were recombined with the Gateway-compatible Tn7 vector pUC18T mini-Tn7T-Gm-GW, resulting in Tn7 constructs pLN4381 (Tn7-katB-HA), pLN4379 (Tn7-katE-HA), and pLN4380 (Tn7-katG-HA), respectively. The Tn7 constructs were coelectroporated with the helper plasmid pTNS2 into their respective mutant strains. The gentamicin-resistant colonies were selected for Tn7 transposition events. To determine the response to H2O2, the resulting strains were grown in liquid M9 Casamino Acids medium at 30°C to an OD600 of 0.3, and H2O2 was added to a final concentration of 1 mM. Samples were taken at the indicated time points and cells harvested and flash frozen in liquid nitrogen. Total protein was extracted by sonication in 1× PBS with complete protease inhibitors and quantified using the Bradford assay. For in planta experiments, the same strains were infiltrated into Arabidopsis leaves at a cell density of 2 × 108 cells ml−1. Leaf disks were sampled with a cork borer at indicated time points after infiltration, and total protein was extracted with 1× PBS with Complete protease inhibitors and quantified using the Bradford assay. The relative levels of catalase proteins were visualized using immunoblotting with anti-HA antibodies.
In planta bacterial growth assay.
Arabidopsis plants were grown at 24°C in a growth chamber under 10-h days. Overnight cultures of bacterial strains grown on KB agar were resuspended to a cell density of 104 or 106 cells ml−1 in 10 mM MgCl2 and infiltrated into fully expanded leaves of 5-week-old Arabidopsis plants with a needleless syringe. Leaf disks of 0.5 cm in diameter were collected with a cork borer at the indicated time points, the samples were ground, and serial dilutions were plated onto KB agar. The bacteria were enumerated after 2 to 3 days at 30°C.
RESULTS
All three DC3000 catalases are active, and KatG is the major housekeeping catalase in culture.
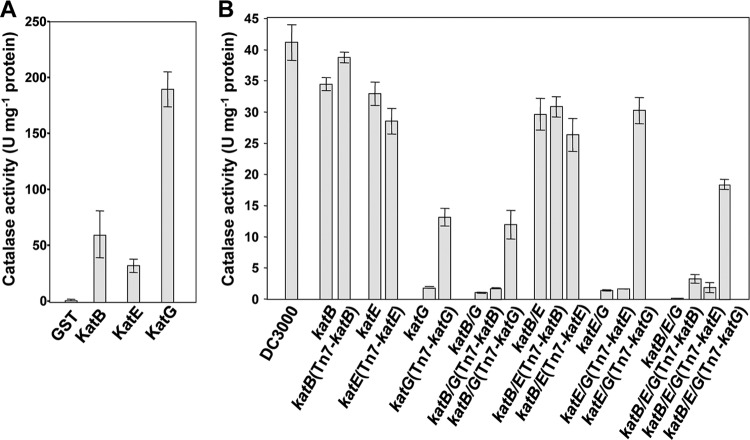
Three catalase genes, katB, katE, and katG, were identified in the DC3000 genome (4). In order to understand the relative importance of these catalases for DC3000, we first determined if these genes encode active catalases. The recombinant catalases were expressed as N-terminal GST fusions in E. coli and purified using glutathione resin. Specific activities of each purified catalase-GST as well as that of a GST control were determined spectrophotometrically by following the loss of H2O2 at A240 over time (42). All three catalases were active, and KatG's in vitro specific activity was about 4-fold higher than those of KatB and KatE (Fig. 1A).
Fig 1.
KatG has the highest specific activity of recombinant DC3000 catalases, and katG mutants have the greatest reduction in catalase activity in in vitro catalase assays. (A) Catalase activity of purified recombinant catalase proteins. The catalases KatB, KatE, and KatG from DC3000 were expressed as N-terminal GST fusions in E. coli. Specific catalase activity was determined for the purified proteins by measuring the rate of H2O2 degradation (reduction of absorbance at 240 nm). One unit of catalase is defined as the amount of enzyme that decomposes 1.0 μmol of H2O2 per minute at pH 7.5. The values are means ± standard errors from six replicates. All three recombinant proteins are active catalases, and KatG has a 4-fold-higher specific activity than KatB and KatE. (B) Catalase activity in crude extracts of DC3000 and catalase mutant strains. Wild-type DC3000, the catalase mutants, and their complemented strains were grown in M9 Casamino Acids liquid medium. The catalase activity of total soluble protein extracts was determined using a fluorometric catalase detection kit. Values are means ± standard errors from six replicates. All strains lacking katG showed significantly less catalase activity than wild-type DC3000 or strains lacking only katB and/or katE. katB/E, katB/G, katE/G, and katB/E/G stand for katB katE, katB katG, and katE katG double mutants and katB katE katG triple mutants, respectively.
As the in vitro activities of recombinant enzymes do not necessarily reflect their relative importance in vivo, we determined the contribution of each catalase to the total catalase activity of DC3000 using mutants. With unmarked mutagenesis (17), we constructed catalase single mutants UNL204 (katB), UNL213 (katE), and UNL222 (katG); double mutants UNL211 (katB katE), UNL223 (katB katG), and UNL225 (katE katG); and a catalase triple mutant, UNL254 (katB katE katG). The total catalase activity of the mutant strains was then compared to that of wild-type DC3000. The strains were grown to stationary phase in liquid Casamino Acids M9 medium, the cells were harvested and lysed, and the total catalase activity of the crude extracts was determined using a catalase fluorometric detection kit. All catalase mutant strains lacking katG were substantially reduced in catalase activity (Fig. 1B). Single or double catalase mutants that lacked katG displayed a 95% reduction in catalase activity compared to that of wild-type DC3000. In contrast, single mutants lacking only katB or katE had catalase activity comparable to that of wild-type DC3000 (Fig. 1B). These data demonstrate that KatG is responsible for the majority of the catalase activity of DC3000 in culture in a manner that is nonredundant with the activities of KatB and KatE. Remarkably, the catalase triple mutant lacking all three catalases was almost completely devoid of measurable catalase activity, indicating that KatB and KatE also contributed a small proportion of the total catalase activity of DC3000 culture (Fig. 1B). Growth curves in rich media were performed for DC3000 and the DC3000 catalase triple mutant, and no significant growth differences were observed (data not shown). This is interesting because it suggests that other enzymes such as peroxidases can protect DC3000 from respiration-related oxidative stress in the absence of these catalases.
To complement the mutants, we expressed catalase genes individually under the control of their native promoters using a Tn7 expression system (7). The resulting strains were grown in culture, and their catalase activity was determined as described above. Complementation with katG significantly increased the catalase activity of all the mutants lacking katG (Fig. 1B), confirming that KatG is the major catalase of DC3000 cultures. Complementation with either katB or katE led to a significant increase of the catalase activity only in the triple mutant (Fig. 1B), indicating that KatB and KatE are indeed functional in DC3000 and probably act in a redundant manner in culture. The 4-fold-higher specific activity of KatG than of KatB and KatE in vitro can partially explain why KatG is the predominant catalase in DC3000 when grown in culture. However, the 95% reduction in total catalase activity of the katG single mutant compared to that of wild-type DC3000 suggests that factors such as expression levels of the enzymes also contribute to predominance of KatG.
KatB accumulation in DC3000 is strongly induced by exogenous H2O2.
KatG appears to be the major catalase of DC3000 in culture; however, KatB and KatE are also functional. It is therefore likely that these catalases play specific roles in DC3000. One possible role is the removal of exogenous H2O2 that would otherwise be toxic to the bacteria. Catalases with this function are often induced by H2O2. Therefore, we monitored the levels of the catalases of DC3000 in response to exogenous H2O2 using a two-pronged approach. First, semiquantitative RT-PCR of DC3000 was used to measure the responsiveness of each catalase to the addition of 1 mM H2O2 (Fig. 2A). Second, immunoblots of catalase single mutants complemented with their respective hemagglutinin-tagged catalases, under the control of their native promoter, were used to determine protein levels of the catalases in culture in response to 1 mM H2O2 (Fig. 2B). In the absence of H2O2, at time zero KatE-HA and KatG-HA were present at similar levels, whereas KatB-HA was below the level of detection (Fig. 2B). Upon addition of H2O2, katB showed a massive increase of transcripts, of around 200-fold, while katG displayed a more modest induction (Fig. 2A). This increased transcription correlated with an induction of KatB-HA at the protein level (Fig. 2B). The levels of KatB and KatE were reduced at about 60 min, presumably as H2O2 is removed by the catalases (Fig. 2B). KatG showed a slight increase in accumulation at around 120 min that potentially correlates with a reduction in the amount of KatB and KatE. The low levels of KatB in culture and its rapid and massive accumulation in response to exogenous H2O2 strongly suggest that a major function of this catalase in DC3000 is the detoxification of exogenous H2O2.
Fig 2.
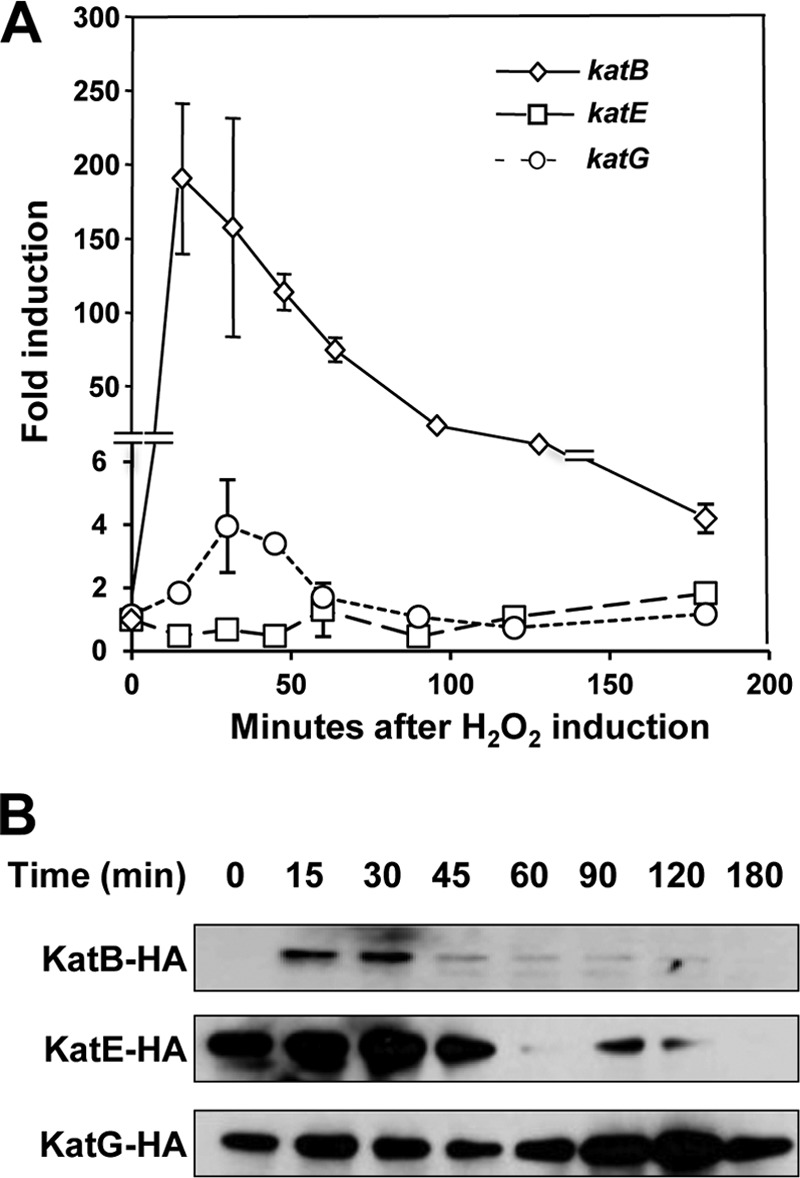
Induction of DC3000 catalases in response to H2O2. (A) Expression of DC3000 catalase genes as determined by semiquantitative RT-PCR. Wild-type DC3000 was grown in M9 Casamino Acids medium to an OD600 of 0.2, and H2O2 was added to a final concentration of 1 mM. Catalase gene expression relative to 16S rRNA was determined at the indicated time points by semiquantitative RT-PCR, and fold induction in response to H2O2 was calculated. Values are means of fold increase ± standard deviations from three replicates of each gene compared to itself in an uninduced control. The expression of katB is rapidly and substantially increased after H2O2 addition. (B) Accumulation of catalase proteins detected with immunoblots. Catalase single mutants complemented with HA-tagged catalases driven by their native promoters in a Tn7 system were grown in M9 Casamino Acids medium to an OD600 of 0.2, and H2O2 was added to a final concentration of 1 mM. Catalase protein levels were determined at the indicated time points by immunoblotting with anti-HA antibodies. KatE and KatG are present at higher levels than KatB, but KatB's protein levels are more affected by H2O2. The experiments were repeated three times with similar results.
The catalases KatB and KatG protect DC3000 from exogenous and endogenous H2O2 toxicity.
The expression patterns of DC3000 catalases indicate that KatB may play a major role in detoxifying exogenous H2O2 (Fig. 2). To investigate this, we performed H2O2 inhibition zone assays (43, 49) with the catalase mutant strains. In this assay, bacteria are plated on solid KB media and H2O2 is spotted onto a filter paper disk placed in the center of the plate. H2O2 diffuses through the media, and the area of growth inhibition observed correlates with the concentration of H2O2 that the bacterial strains are able to detoxify. The smaller the inhibition zone, the greater the ability of the bacteria to detoxify H2O2.
As expected, of the strains tested, the catalase triple mutant was the most sensitive to H2O2, exhibiting the largest growth inhibition zone (Fig. 3A). This is consistent with its almost complete loss of catalase activity (Fig. 1B). We next determined whether individual catalases were able to increase the resistance of the triple mutant to H2O2. This was accomplished using the triple mutant strain complemented with single catalases under the control of their native promoters via the Tn7 system. This assay showed that the expression of katB in the triple mutant led to the most prominent reduction in the size of the H2O2 inhibition zone (Fig. 3A). The expression of katG in the triple mutant also enhanced its resistance to H2O2, although to a lesser extent than katB (Fig. 3A). However, the expression of katE in the triple mutant did not significantly alter its ability to detoxify H2O2 (Fig. 3A). These data show that both KatB and KatG enhanced the H2O2 tolerance of DC3000, allowing it to grow better in the presence of exogenous H2O2.
Fig 3.
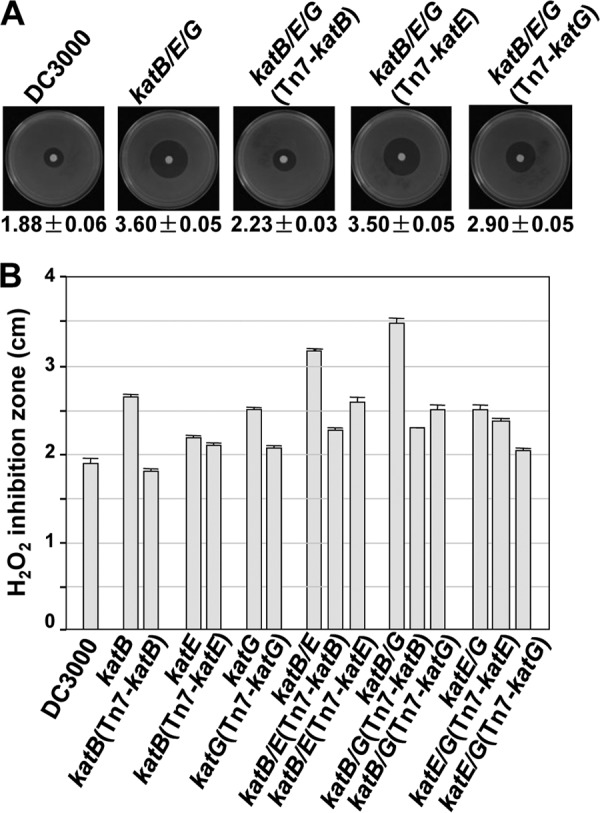
H2O2 inhibition zone assays. (A) H2O2 inhibition zone assay for DC3000, the catalase triple mutant, and its complemented strains. Representative pictures of the strains tested are shown. Below each picture is the diameter (cm) of the H2O2 inhibition zone ± standard deviation from three replicates. The catalase triple mutant is hypersensitive to H2O2. Complementation with KatB and KatG but not KatE significantly increased the tolerance of the catalase triple mutant to H2O2. (B) H2O2 inhibition zones for wild-type DC3000, the catalase single and double mutants, and their complemented strains ± standard deviations of three replicates. The loss of KatB or KatG resulted in an increased sensitivity to H2O2 in an additive fashion, indicating that these two catalases have nonredundant roles in protecting DC3000 from exogenous H2O2. The experiments were repeated four times with similar results.
To confirm the relative importance of KatB and KatG in H2O2 detoxification, we examined the H2O2 inhibition zones of the single and double catalase mutants and their complemented strains. Of the catalase single mutants, the katB mutant was most sensitive to H2O2, followed by the katG mutant, while the katE mutant displayed an almost wild-type ability to detoxify H2O2 (Fig. 3B). The phenotypes of the katB and katG mutants were complemented by katB and katG, respectively. Of the catalase double mutants, the katB katG mutant had the largest inhibition zone, followed by the katB katE and katE katG mutants, respectively (Fig. 3B). Tn7 complementation with either katB or katG reduced the inhibition zones of the double mutants lacking either katB or katG, respectively (Fig. 3B). The additive effect of the loss of katB and katG on the sensitivity of DC3000 to H2O2 in the double mutant was expected. Concurrent with the minor contribution of katE, this double mutant is as sensitive to H2O2 as the catalase triple mutant. An unexpected result was the additive effect of the loss of katE to the loss of katB in the katB katE double mutant, which was not seen in the katE katG double mutant (Fig. 3B). These data suggest that KatE may play a subtle role in detoxifying H2O2 that is linked to that of KatB, possibly due to common regulatory factors. These data demonstrate that KatB and KatG play nonredundant roles in DC3000 for the detoxification of exogenous H2O2.
The removal of exogenous H2O2 is not the only way that catalases can protect the cell. H2O2 is also produced endogenously as a by-product from many intracellular reactions. Some catalases can act as scavengers to prevent the accumulation of toxic levels of intracellular H2O2. To determine if KatB, KatE, or KatG of DC3000 is involved in the removal of endogenous H2O2, we performed menadione growth inhibition assays. Menadione is often used to mimic intracellular H2O2 stress in bacteria because it generates an endogenous source of H2O2 (11, 13, 31, 35). To investigate the ability of catalases to remove endogenous H2O2, we compared the growth of wild-type DC3000 to that of the catalase mutants on plates containing menadione. Of the single mutant strains, only the katG mutant displayed significantly reduced growth on menadione compared to that of DC3000 (Fig. 4A). Complementation of the katG mutant restored its growth to wild-type levels, suggesting that KatG is the DC3000 catalase responsible for detoxifying the majority of H2O2 generated by menadione.
Fig 4.
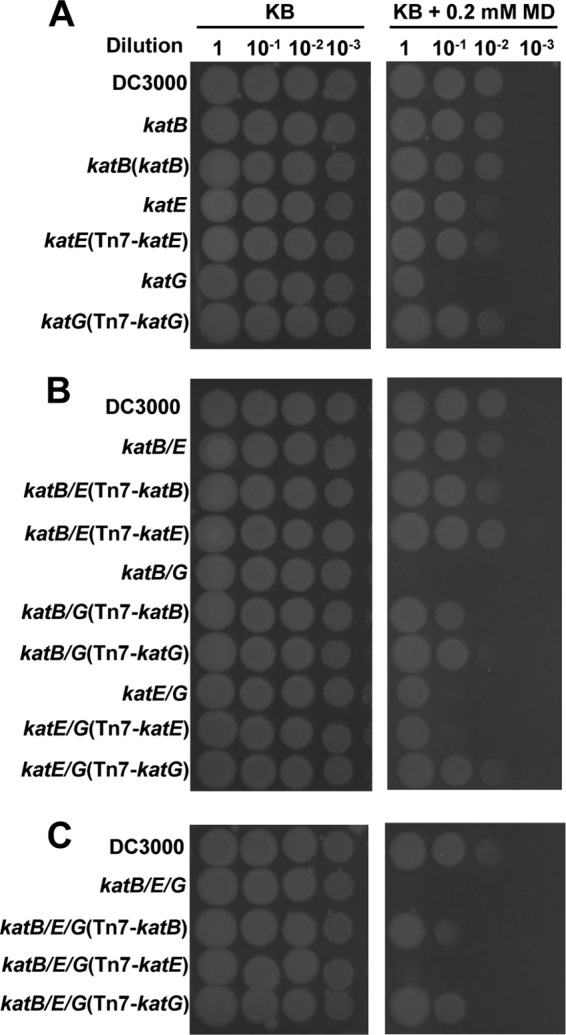
Menadione growth inhibition assays. Wild-type DC3000, the single (A), double (B), and triple (C) catalase mutants, and their Tn7 complemented strains were assessed for their sensitivity to menadione toxicity. The bacteria were resuspended to 1 × 108 cells ml−1, and 10 μl of 10-fold serial dilutions was spotted on KB plates with or without 0.2 mM menadione (MD). Pictures were taken 48 h later. The pictures are representative of three independent experiments.
Interestingly, the katB katG double mutant and the triple mutant were further impaired in their ability to grow on menadione-containing plates (Fig. 4B). Expression of either katB or katG in the katB katG double mutant or the triple mutant enhanced their menadione tolerance, indicating that both KatB and KatG function to protect cells from menadione toxicity (Fig. 4B and C). In contrast, disruption of katE in the single, double, or triple mutants had little effect on bacterial growth, nor was the expression of katE able to exert a protective effect on any of the mutants, suggesting that KatE is not able to alleviate menadione toxicity. These data clearly show that in DC3000 KatG is responsible for removing the majority of the menadione-generated (i.e., intracellular) H2O2, while KatB removes a minor portion of it. KatE is apparently not significantly involved in removing this source of endogenous H2O2.
All three catalases are induced in planta, and KatB and KatG have nonredundant roles in the virulence of DC3000.
Plants initiate a series of immune responses when perceiving microorganisms. One of the earliest responses is an oxidative burst that includes the production of H2O2 by plasma membrane-localized NADPH oxidases and peroxidases (10, 36, 47). The resulting rapid increase in apoplastic ROS is potentially toxic for DC3000. As catalases are responsible for protecting DC3000 from exogenous H2O2, we speculated they may also provide protection against this oxidative burst and, therefore, be important for the virulence of DC3000. To determine the roles of these catalases in the virulence of DC3000, we first analyzed whether they accumulate when the bacteria infect their host plants. Semiquantitative RT-PCR was used to measure the expression of each catalase in wild-type DC3000 syringe infiltrated at 2 ×108 cells ml−1 into the leaves of Arabidopsis plants (Fig. 5A). All three catalases were transiently induced in planta between 0.5 and 2 h postinoculation, with katB and katE showing a 10-fold induction and katG a 4-fold induction (Fig. 5A). Protein levels of the catalases in planta were assessed using anti-HA immunoblots of Arabidopsis leaves infiltrated with 2 ×108 cells ml−1 of the catalase single mutants complemented with their respective HA-tagged catalases under the control of their native promoters (Fig. 5B). Immunoblots showed that KatE and KatG accumulated to similar levels but KatB expression was below the level of detection (Fig. 5B). These data show that the transcripts of all three catalases accumulate in DC3000 during its interaction with Arabidopsis. It is likely that they are important for P. syringae to survive in its host.
Fig 5.
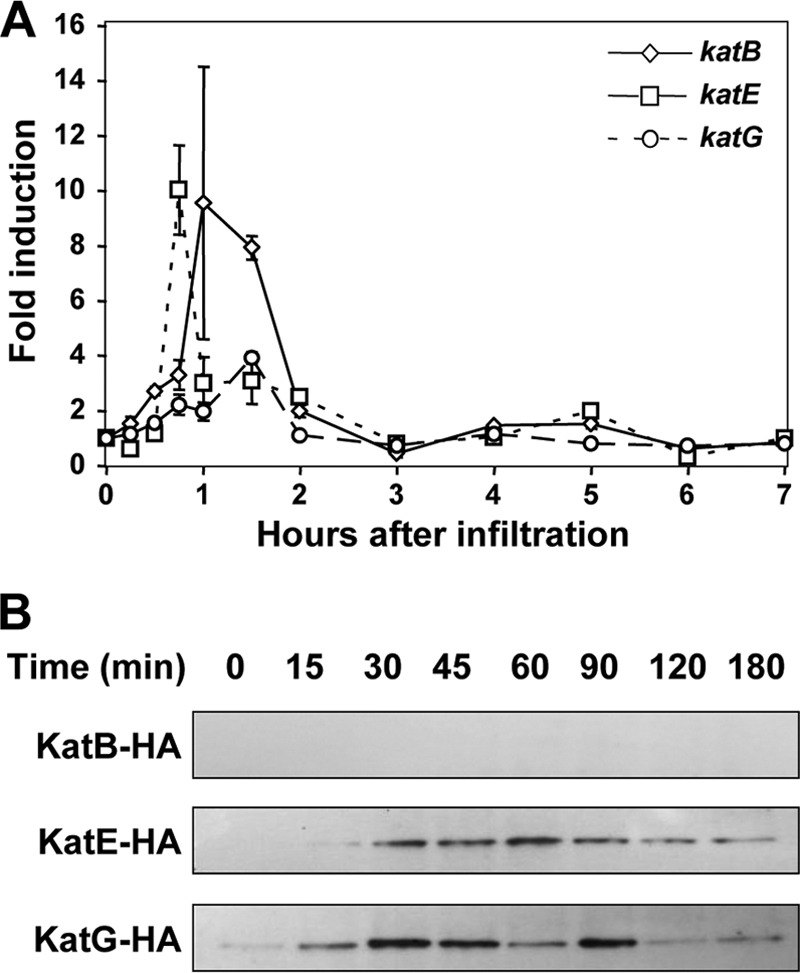
Induction of DC3000 catalases in planta. (A) Transcription of DC3000 catalase genes in planta. Wild-type DC3000 at 2 × 108 cells ml−1 was infiltrated into Arabidopsis leaves. Tissues were harvested at the indicated times, and catalase gene expression relative to 16S rRNA was determined by semiquantitative RT-PCR. Values are means of fold increase ± standard deviations from three replicates of each gene compared to itself in an uninduced control. The expression of all three catalases was induced in planta. (B) Accumulation of DC3000 catalase proteins in planta. Catalase single mutant strains expressing HA-tagged versions of their respective catalases driven by their native promoters in a Tn7 system were syringe infiltrated at 2 × 108 cells ml−1 into Arabidopsis leaves. Tissues were harvested at the indicated times, and catalase protein levels were determined by immunoblotting with anti-HA antibodies. The catalases KatE and KatG are present at higher levels than KatB and induced when DC3000 is in planta. Each experiment was repeated three times with similar results.
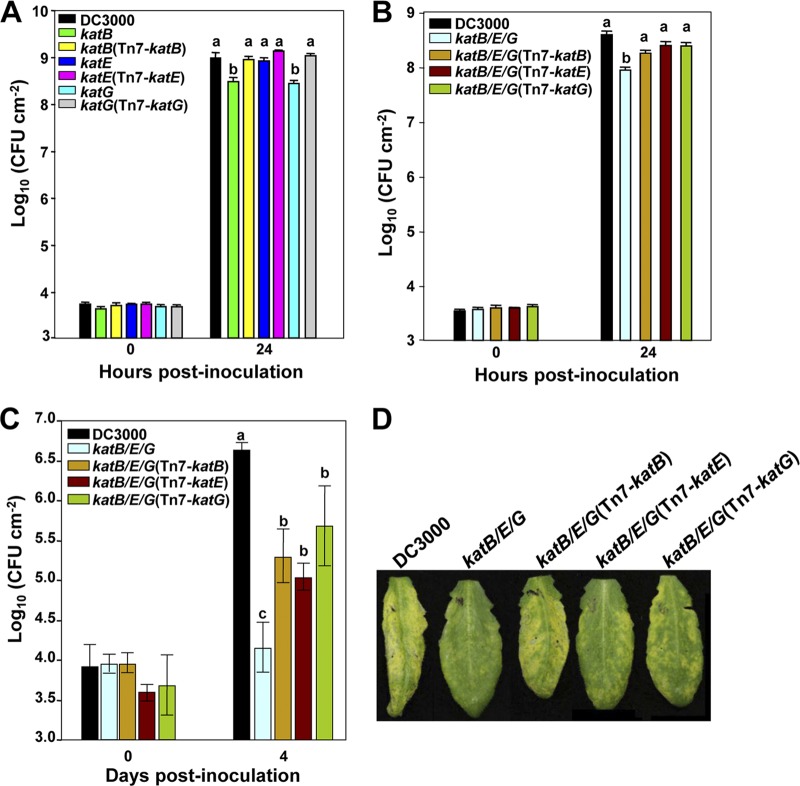
To directly test if these catalases impact the virulence of DC3000, we compared the growth of the catalase single mutants and their complemented strains to that of wild-type DC3000 in Arabidopsis. We syringe infiltrated 1 × 106 cells ml−1 of bacteria into Arabidopsis plants and enumerated the bacteria in leaf disks 0 and 24 h after infiltration (Fig. 6A). Both katB and katG single mutants displayed a 3-fold reduction in growth in planta compared to that of wild-type DC3000, and their growth was restored in the complemented mutants. However, the growth of the katE mutant and its complemented strain was similar to that of wild-type DC3000. These data indicate that KatB and KatG play nonredundant roles in the virulence of DC3000.
Fig 6.
Virulence assay for DC3000 catalase mutants. (A) Wild-type DC3000, the catalase single mutants, and their complemented strains were syringe infiltrated at 1 × 106 cells ml−1 into Arabidopsis leaves. Leaf disks were sampled 0 and 24 h postinoculation, and bacteria were enumerated. The katB and katG single mutants but not the katE mutant displayed reduced virulence on Arabidopsis compared to that of DC3000. (B) Wild-type DC3000, the catalase triple mutant, and its complemented strains were syringe infiltrated at 1 × 106 cells ml−1 into Arabidopsis leaves. Bacteria were enumerated in leaf disks sampled 0 and 24 h postinoculation. The triple mutant showed a significantly reduced growth in Arabidopsis compared to that of DC3000 and was partially complemented by all three catalases. (C) Wild-type DC3000, the catalase triple mutant, and its complemented strains were syringe infiltrated at 1 × 104 cells ml−1 into Arabidopsis leaves. Four days later, leaf disks were sampled and bacteria enumerated. The virulence phenotype observed at 4 days is consistent with that observed at 24 h postinoculation. Results are representative of three independent repeats. (D) Symptoms of indicated strains from panel C 4 days after inoculation. Different letters over the bars in panels A through C indicate a significant difference (P ≤ 0.05). All the experiments were repeated three times with similar results.
We next determined the ability of each catalase to enhance the in planta growth of a DC3000 mutant lacking all three catalases. To accomplish this, we compared the growth of DC3000 to that of the catalase triple mutant and the catalase triple mutant complemented with each catalase under the control of its native promoter. We monitored the number of bacteria in Arabidopsis leaf disks at 0 and 24 h after syringe infiltration (Fig. 6B). As expected, the catalase triple mutant was significantly impaired in its ability to grow in planta compared to wild-type DC3000, probably due to an inability to remove H2O2 produced during the oxidative burst (Fig. 6B). The triple mutant showed 5-fold-lower growth than DC3000 at 24 h after inoculation. Surprisingly, all three catalases were able to enhance the growth of the triple mutant in planta to similar extents, although none was able to fully restore its growth to wild-type levels.
As the oxidative burst is an early response of plants to microbial infection, the reduction of the growth of the catalase triple mutant at 24 h after inoculation is in line with it being compromised in its ability to overcome this immune response. We next determined whether the loss of catalase activity resulted in a long-term effect on the virulence of DC3000. To do this, we examined growth of wild-type DC3000, the catalase triple mutant, and its complemented strains at later stages of the infection. We enumerated the bacteria at 4 days after inoculation in Arabidopsis plants syringe infiltrated with bacteria at 1 × 104 cells ml−1. As at the early stages of infection, the growth of the catalase triple mutant was significantly reduced in planta compared to that of wild-type DC3000 at 4 days postinoculation (Fig. 6C). In line with its reduced virulence, the catalase triple mutant was also reduced in symptom development 4 days after inoculation compared with DC3000 (Fig. 6D). Consistently, the complementation of the triple mutant with single catalases was able to partially rescue its in planta growth and disease symptoms (Fig. 6C and D). Complementation of the triple mutant with katB, katE, or katG gives strains growth reductions of 12-, 13-, and 10-fold, respectively, compared to the level for DC3000 at 4 days after inoculation. Growth of the single mutants at 4 days after inoculation was also measured. Both katB and katG mutants showed an 11-fold growth reduction compared to the level for DC3000 (data not shown). These data suggest that KatB, KatE, and KatG are all able to partially protect DC3000 from the toxic effects of plant-derived H2O2, allowing it to survive and multiply in planta. However, the reduced virulence of the katB and katG single mutants indicates that these two catalases act as virulence factors in a nonredundant manner.
DISCUSSION
In this study, we demonstrated that all three DC3000 catalases (KatB, KatE, and KatG) are active in vitro as purified recombinant enzymes and that KatG has about four times the specific activity of KatB or KatE (Fig. 1A). Furthermore, we made DC3000 single- and polycatalase mutants and found that mutants that lacked katG were the most reduced in total catalase activity in assays that measured the catalase activity present in soluble extracts (Fig. 1B). Interestingly, expression of katB was induced substantially more than that of katE and katG after exposure to H2O2 (Fig. 2A), and KatB-HA was clearly induced by H2O2 more substantially than KatE-HA or KatG-HA (Fig. 2B). Based on data shown in Fig. 1 and 2, it appears that KatG represents a housekeeping catalase while the other two catalases, particularly KatB, are involved in oxidative stress conditions. This is further supported by results from the H2O2 inhibition assays that indicated that the DC3000 polycatalase mutant growth was restored the most when katB was reintroduced into this mutant (Fig. 3A). In the H2O2 inhibition assays using the different combinations of the DC3000 single and double catalase mutants, it appeared that KatB and KatG contributed the most to H2O2 protection (Fig. 3B). However, KatE did partially complement the DC3000 double mutants, suggesting that it also contributed to protection from extracellular H2O2.
The menadione growth inhibition assays on the single catalase mutants clearly indicate that KatG plays the most important role in detoxifying intracellular H2O2 (Fig. 4A).The assays using polycatalase mutants showed that KatB contributed to detoxifying intracellular H2O2 whereas KatE did not appear to contribute to H2O2 detoxification (Fig. 4B and C). Collectively, our results show that KatB and KatG are the catalases of DC3000 that are primarily responsible for detoxifying H2O2. However, the two catalases have differing roles depending on the origin of the H2O2. In the case of exogenous H2O2, KatB and KatG appear to have an additive effect, with KatB providing slightly more protection than KatG. In the case of endogenous H2O2, KatG appears to provide the majority of the protection and KatB's contribution is significant only in the absence of KatG.
The apparent minor role of KatE in DC3000 observed in our assays may be due to it functioning in hitherto unknown conditions not examined in this study. For instance, KatC, a KatE homolog in P. aeruginosa, is a temperature-dependent catalase that protects the bacteria from H2O2 at 42°C but not at 30°C or 37°C (32). Thus, it is possible that KatE is important only under specific environmental conditions. A similar pattern of catalase function is seen in X. campestris pv. campestris, a pathogen that has two monofunctional catalases, KatA and KatE, and a bifunctional catalase, KatG (21). In our studies, DC3000 KatE does provide a small amount of catalase activity, as the katB katG double mutant has higher catalase activity than the triple mutant (Fig. 1B).
It is not an uncommon survival strategy for pathogenic bacteria to utilize both a housekeeping catalase and an H2O2-inducible catalase when they are at risk of exposure to high levels of host-derived H2O2. Indeed, such a strategy is seen in the opportunistic human pathogen P. aeruginosa, which has three monofunctional catalases, KatA, KatB, and KatC. KatA is the housekeeping catalase, and KatB is expressed at a high level only upon exposure to H2O2 or an oxidative-stress-generating compound (3, 34). In P. aeruginosa, KatA is present in the cytoplasm and periplasm while KatB is purely cytoplasmic (3). Several lines of evidence indicate that the opposite is true for KatB and KatG in DC3000. KatB from DC3000 is 95% identical at the amino acid level to the catalase CatF from P. syringae pv. syringae 61 (28), which is located in both the cytoplasm and the periplasm (27). Furthermore, the signal peptide prediction programs SignalP (http://www.cbs.dtu.dk/services/SignalP/) and SIG-Pred (http://bmbpcu36.leeds.ac.uk/prot_analysis/Signal.html) indicate that the N-terminal 26 amino acids of the DC3000 KatB protein likely acts as a signal peptide, while neither KatE nor KatG has predicted signal peptides. These data indicate that KatB of DC3000 is likely in the periplasm and cytoplasm, while KatE and KatG are likely cytoplasmically localized.
The respective roles of KatB and KatG in DC3000 in response to menadione toxicity differ from the response to exogenous H2O2 toxicity. Menadione is a redox-cycling quinone that can reduce oxygen to superoxide, which can then be converted to H2O2 by superoxide dismutase (35). KatG from DC3000 may provide a higher level of protection against menadione than KatB due to its predicted predominantly cytoplasmic location, which would place it in closer proximity to the endogenous H2O2 generated by menadione. An alternate possibility is that the bifunctional activity of KatG enhances its ability to detoxify menadione. Other bifunctional catalases have been shown to play a major role in detoxifying menadione, including KatG from X. campestris pv. campestris strain ATCC 33913 (21) and KatA (a homolog of KatG) from A. tumefaciens NTL4 (35). It is possible that the enhanced ability of bifunctional catalases to detoxify menadione arises from an ability to remove organic hydroperoxides generated by menadione. However, KatA has been shown not to play a major role in protecting A. tumefaciens from organic hydroperoxide toxicity (35). These data suggest that bifunctional catalases such as KatG may be particularly effective at relieving menadione toxicity, but their mechanism for doing so remains, as yet, undetermined.
The expression of all three catalases of DC3000 is induced when the bacteria are in planta (Fig. 5), suggesting that their activity is required in this environment. Single mutants in either katB or katG have reduced growth in planta compared to wild-type DC3000 (Fig. 6A), indicating that these catalases play nonredundant roles in the virulence of DC3000 (Fig. 6). The catalase triple mutant was severely impaired in its ability to grow and cause disease in Arabidopsis, and all three catalases enhanced the growth in planta of the triple mutant when separately reintroduced, indicating that they each provide a degree of protection against ROS stress during pathogenesis (Fig. 6B and C). These data suggest that KatB, KatE, and KatG can all contribute to P. syringae pathogenesis, yet they are individually incapable of restoring wild-type growth. This nonredundant role of catalases in virulence is also seen in X. campestris pv. campestris, in which both katA and katG single mutants are avirulent in its host Chinese radish (Raphanus sativus) (21). Catalases have also been shown to be important in the virulence of other plant-pathogenic bacteria. For example, an A. tumefaciens mutant with loss of function in its bifunctional catalase, katA, has reduced tumor development (49), and an X. axonopodis pv. citri mutant in its monofunctional catalase, katE, has increased sensitivity to H2O2 toxicity and reduced virulence in citrus plants (45). The differential use of catalases by many plant pathogens to achieve the same goal lends support to the idea that these catalases were independently adapted to allow the bacteria to overcome defensive production of H2O2 by plants.
The more general importance of catalases for bacterium-plant interactions is seen in their use by symbiotic bacteria such as Mesorhizobium loti, in which the disruption of a single catalase, katE, led to decreased nitrogen fixation (15). The symbiotic bacterium Sinorhizobium meliloti is similar to DC3000, as it has three catalases, two monofunctional catalases (KatA and KatC), and one bifunctional catalase-peroxidase (KatB). KatA and KatB are both periplasmic, and katA but not katB is inducible by H2O2 (16). The induction of katC is observed only under heat stress or salt stress or upon treatment with ethanol or the ROS generator paraquat (41). S. meliloti catalase single mutants are unaltered in their ability to fix nitrogen or form nodules in their host plant, but both katA katC and katB katC double mutants show reduced nitrogen fixation and nodule-forming abilities (20, 41). Interestingly, the double mutants interfere with nodule formation at different steps, indicating that katA and katB function at different stages of the interaction of the rhizobia with the plants (20). Due to the similarities with DC3000, this suggests that KatB and KatG from DC3000 may aid the bacteria's growth at different stages of the infection process. Future studies will likely clarify how KatB and KatG enable DC3000 to be a successful pathogen. From the data in our study, one could hypothesize that KatB may be providing direct protection against the oxidative burst while KatG may aid in the detoxification of ROS-generating compounds or the removal of H2O2 produced by stress on the bacteria's metabolism.
When a microorganism comes in contact with a plant cell, the plant's immune response is activated by PTI or effector-triggered immunity, resulting in a wide array of immune responses, including responses that are directed toward fungal, viral, or bacterial pathogens (33, 44). Thus, while it has been known for many years that these immune responses are successful against saprophytic bacteria and nonadapted and avirulent pathogens, it is not clear which specific immune responses are successful at inhibiting or killing bacteria in the apoplast. When PTI is induced, the ability of P. syringae to inject type III effectors is restricted within the first hour after induction (8), indicating that early immune responses are effective against P. syringae. ROS production occurs within the first few minutes after PTI is induced (48), suggesting that it may be an important component of the plant immune response to bacteria. The contribution of the catalases of P. syringae to virulence further suggests this to be the case.
ACKNOWLEDGMENTS
We thank the Alfano research group members for fruitful discussions regarding the experiments described in this article. We also thank Wenjing Guan, Danellie Fortune, and Alexis Paspalof for technical assistance.
This research was supported by grants from the U.S. Department of Agriculture (award no. 2007-35319-18336) and the National Institutes of Health (award no. 1R01AI069146-01A2 and P20RR-017675) and funds from the Center for Plant Science Innovation at the University of Nebraska.
Footnotes
Published ahead of print 13 July 2012
REFERENCES
- 1. Block A, Alfano JR. 2011. Plant targets for Pseudomonas syringae type III effectors: virulence targets or guarded decoys? Curr. Opin. Microbiol. 14:39–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boller T, Felix G. 2009. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60:379–406 [DOI] [PubMed] [Google Scholar]
- 3. Brown SM, Howell ML, Vasil ML, Anderson AJ, Hassett DJ. 1995. Cloning and characterization of the katB gene of Pseudomonas aeruginosa encoding a hydrogen peroxide-inducible catalase: purification of KatB, cellular localization, and demonstration that it is essential for optimal resistance to hydrogen peroxide. J. Bacteriol. 177:6536–6544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Buell CR, et al. 2003. The complete sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. U. S. A. 100:10181–10186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cabiscol E, Tamarit J, Ros J. 2000. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int. Microbiol. 3:3–8 [PubMed] [Google Scholar]
- 6. Chaouch S, Queval G, Noctor G. 2012. AtRbohF is a crucial modulator of defence-associated metabolism and a key actor in the interplay between intracellular oxidative stress and pathogenesis responses in Arabidopsis. Plant J. 69:613–627 [DOI] [PubMed] [Google Scholar]
- 7. Choi KH, et al. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2:443–448 [DOI] [PubMed] [Google Scholar]
- 8. Crabill E, Joe A, Block A, van Rooyen JM, Alfano JR. 2010. Plant immunity directly or indirectly restricts the injection of type III effectors by the Pseudomonas syringae type III secretion system. Plant Physiol. 154:233–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cuppels DA. 1986. Generation and characterization of Tn5 insertion mutations in Pseudomonas syringae pv. tomato. Appl. Environ. Microbiol. 51:323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Daudi A, et al. 2012. The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. Plant Cell 24:275–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Farr SB, Kogoma T. 1991. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol. Rev. 55:561–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Feil H, et al. 2005. Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proc. Natl. Acad. Sci. U. S. A. 102:11064–11069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Flattery-O'Brien J, Collinson LP, Dawes IW. 1993. Saccharomyces cerevisiae has an inducible response to menadione which differs from that to hydrogen peroxide. J. Gen. Microbiol. 139:501–507 [DOI] [PubMed] [Google Scholar]
- 14. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580 [DOI] [PubMed] [Google Scholar]
- 15. Hanyu M, Fujimoto H, Tejima K, Saeki K. 2009. Functional differences of two distinct catalases in Mesorhizobium loti MAFF303099 under free-living and symbiotic conditions. J. Bacteriol. 191:1463–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Herouart D, et al. 1996. Cloning and characterization of the katA gene of Rhizobium meliloti encoding a hydrogen peroxide-inducible catalase. J. Bacteriol. 178:6802–6809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. House BL, Mortimer MW, Kahn ML. 2004. New recombination methods for Sinorhizobium meliloti genetics. Appl. Environ. Microbiol. 70:2806–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Imlay JA, Linn S. 1988. DNA damage and oxygen radical toxicity. Science 240:1302–1309 [DOI] [PubMed] [Google Scholar]
- 19. Jamet A, Kiss E, Batut J, Puppo A, Herouart D. 2005. The katA catalase gene is regulated by OxyR in both free-living and symbiotic Sinorhizobium meliloti. J. Bacteriol. 187:376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jamet A, Sigaud S, Van de Sype G, Puppo A, Herouart D. 2003. Expression of the bacterial catalase genes during Sinorhizobium meliloti-Medicago sativa symbiosis and their crucial role during the infection process. Mol. Plant Microbe Interact. 16:217–225 [DOI] [PubMed] [Google Scholar]
- 21. Jittawuttipoka T, Buranajitpakorn S, Vattanaviboon P, Mongkolsuk S. 2009. The catalase-peroxidase KatG is required for virulence of Xanthomonas campestris pv. campestris in a host plant by providing protection against low levels of H2O2. J. Bacteriol. 191:7372–7377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Joardar V, et al. 2005. Whole-genome sequence analysis of Pseudomonas syringae pv. phaseolicola 1448A reveals divergence among pathovars in genes involved in virulence and transposition. J. Bacteriol. 187:6488–6498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Karpinski S, Gabrys H, Mateo A, Karpinska B, Mullineaux PM. 2003. Light perception in plant disease defence signalling. Curr. Opin. Plant Biol. 6:390–396 [DOI] [PubMed] [Google Scholar]
- 24. Katsuwon J, Anderson AJ. 1989. Response of plant-colonizing pseudomonads to hydrogen peroxide. Appl. Environ. Microbiol. 55:2985–2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Keppler LD, Baker CJ. 1989. O-2−-initiated lipid-peroxidation in a bacteria-induced hypersensitive reaction in tobacco cell-suspensions. Phytopathology 79:555–562 [Google Scholar]
- 26. King EO, Ward MK, Raney DE. 1954. Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Med. 22:301–307 [PubMed] [Google Scholar]
- 27. Klotz MG, Hutcheson SW. 1992. Multiple periplasmic catalases in phytopathogenic strains of Pseudomonas syringae. Appl. Environ. Microbiol. 58:2468–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Klotz MG, Kim YC, Katsuwon J, Anderson AJ. 1995. Cloning, characterization and phenotypic expression in Escherichia coli of catF, which encodes the catalytic subunit of catalase isozyme CatF of Pseudomonas syringae. Appl. Microbiol. Biotechnol. 43:656–666 [DOI] [PubMed] [Google Scholar]
- 29. Kwak JM, et al. 2003. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22:2623–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lamb C, Dixon RA. 1997. The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48:251–275 [DOI] [PubMed] [Google Scholar]
- 31. Mongkolsuk S, et al. 1996. Heterologous growth phase- and temperature-dependent expression and H2O2 toxicity protection of a superoxide-inducible monofunctional catalase gene from Xanthomonas oryzae pv. oryzae. J. Bacteriol. 178:3578–3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mossialos D, Tavankar GR, Zlosnik JE, Williams HD. 2006. Defects in a quinol oxidase lead to loss of KatC catalase activity in Pseudomonas aeruginosa: KatC activity is temperature dependent and it requires an intact disulphide bond formation system. Biochem. Biophys. Res. Commun. 341:697–702 [DOI] [PubMed] [Google Scholar]
- 33. Navarro L, et al. 2004. The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 135:1113–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ochsner UA, Vasil ML, Alsabbagh E, Parvatiyar K, Hassett DJ. 2000. Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J. Bacteriol. 182:4533–4544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Prapagdee B, Vattanaviboon P, Mongkolsuk S. 2004. The role of a bifunctional catalase-peroxidase KatA in protection of Agrobacterium tumefaciens from menadione toxicity. FEMS Microbiol. Lett. 232:217–223 [DOI] [PubMed] [Google Scholar]
- 36. Rojas CM, et al. 2012. Glycolate oxidase modulates reactive oxygen species-mediated signal transduction during nonhost resistance in Nicotiana benthamiana and Arabidopsis. Plant Cell 24:336–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rudolph K, Stahmann MA. 1964. Interactions of peroxidases and catalases between Phaseolus vulgaris and Pseudomonas phaseolicola (halo blight of bean). Nature 204:474–475 [DOI] [PubMed] [Google Scholar]
- 38. Ruvkun GB, Ausubel FM. 1981. A general method for site-directed mutagenesis in prokaryotes. Nature 289:85–88 [DOI] [PubMed] [Google Scholar]
- 39. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 40. Segonzac C, Zipfel C. 2011. Activation of plant pattern-recognition receptors by bacteria. Curr. Opin. Microbiol. 14:54–61 [DOI] [PubMed] [Google Scholar]
- 41. Sigaud S, Becquet V, Frendo P, Puppo A, Herouart D. 1999. Differential regulation of two divergent Sinorhizobium meliloti genes for HPII-like catalases during free-living growth and protective role of both catalases during symbiosis. J. Bacteriol. 181:2634–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Singh R, et al. 2008. Comparative study of catalase-peroxidases (KatGs). Arch. Biochem. Biophys. 471:207–214 [DOI] [PubMed] [Google Scholar]
- 43. Subramoni S, Sonti RV. 2005. Growth deficiency of a Xanthomonas oryzae pv. oryzae fur mutant in rice leaves is rescued by ascorbic acid supplementation. Mol. Plant Microbe Interact. 18:644–651 [DOI] [PubMed] [Google Scholar]
- 44. Tao Y, et al. 2003. Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15:317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tondo ML, Petrocelli S, Ottado J, Orellano EG. 2010. The monofunctional catalase KatE of Xanthomonas axonopodis pv. citri is required for full virulence in citrus plants. PLoS One 5:e10803 doi:10.1371/journal.pone.0010803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Torres MA. 2010. ROS in biotic interactions. Physiol. Plant. 138:414–429 [DOI] [PubMed] [Google Scholar]
- 47. Torres MA, Jones JD, Dangl JL. 2005. Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat. Genet. 37:1130–1134 [DOI] [PubMed] [Google Scholar]
- 48. Torres MA, Jones JD, Dangl JL. 2006. Reactive oxygen species signaling in response to pathogens. Plant Physiol. 141:373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xu XQ, Pan SQ. 2000. An Agrobacterium catalase is a virulence factor involved in tumorigenesis. Mol. Microbiol. 35:407–414 [DOI] [PubMed] [Google Scholar]
- 50. Zhang J, et al. 2007. A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1:175–185 [DOI] [PubMed] [Google Scholar]