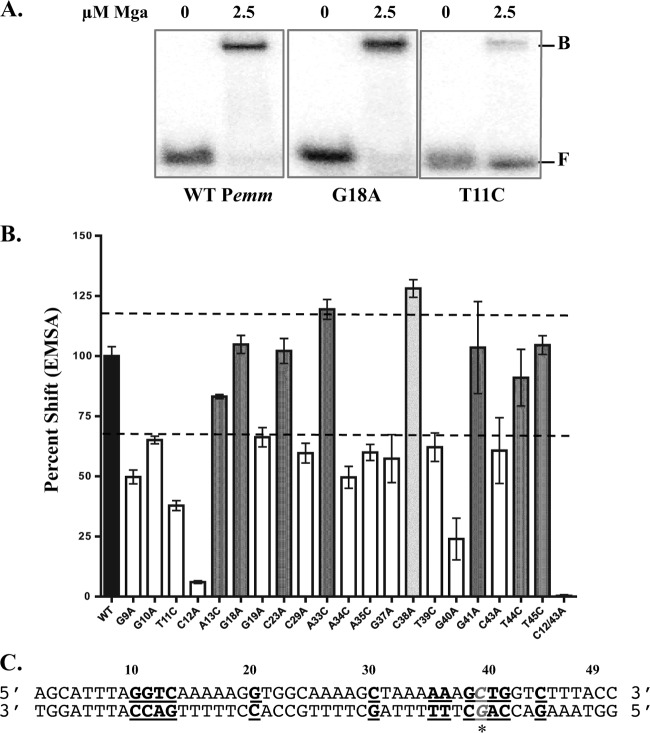
Fig 5.
DNA-binding activity of Pemm site-directed mutations. EMSA analysis of strains with Pemm1 49-mer point mutations compared to the wild-type strain was performed to identify nucleotides important for Mga binding. Each probe (0.1 nM) was incubated with 2.5 μM Mga1-His6 (except the C38A 49-mer was incubated with 1.25 μM Mga1-His6). (A) Representative EMSA results for Pemm1 wild-type (WT), G18A, and T11C 49-mer probes with Mga1-His6 (2.5 μM) or without Mga1-His6 (0) are shown. The positions of free (F) and bound (B) bands are indicated to the right of the gel. (B) Quantification of EMSA results for each Pemm1 49-mer point mutant (shown below the bars) as determined by densitometry and shown as percent shifted by Mga1-His6 compared to wild-type Pemm1. Mutants that shift less than the wild type (white), comparable to the wild type (dark gray), and greater than the wild type (light gray) are indicated. The broken line denotes 70% and 120% of wild-type binding. (C) Schematic diagram showing the locations of all nucleotides within the Pemm1 49-mer identified as important for DNA binding (bold and underlined). The C38A mutation leading to increased binding is shown in gray italic underlined letters with an asterisk.