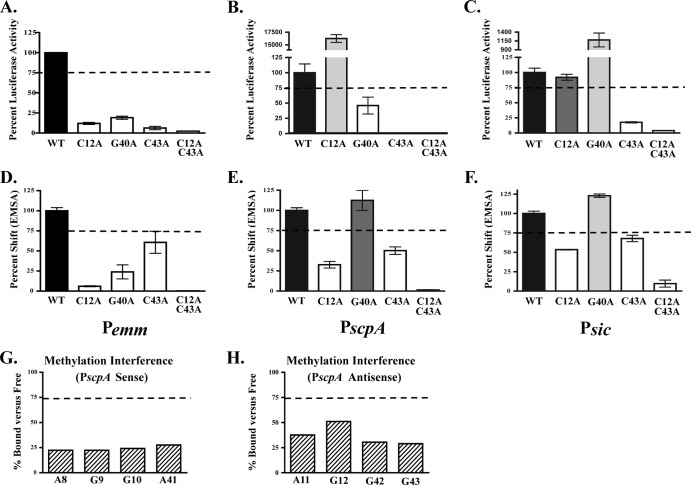
Fig 6.
Role of functional Pemm nucleotides conserved in PscpA and Psic. Three nucleotides conserved in Pemm, PscpA, and Psic (C12, G40, and C43) and found to be important for both activity and DNA binding in Pemm1 were chosen for analysis. (A to F) Single mutations (C12A, G40A, and C43A) and the C12A C43A double mutation in Pemm (A), PscpA (B), and Psic (C) were assayed for promoter activity by luciferase reporter assay (A to C) and DNA binding by EMSA (D to F) for each promoter. Quantification of RLU is shown as percent luciferase activity compared to the respective wild-type promoter. Mutants showing less than 75% wild-type activity (light gray bars), 75 to 100% of wild-type activity (dark gray bars), and greater than wild-type activity (white bars) are indicated. Quantification of EMSA Pemm (D), PscpA (E), and Psic (F) is shown as percent shifted by Mga1-His6 compared to the respective wild type. Mutants that shift less than the wild type (light gray bars), comparable to the wild type (dark gray bars), and greater than the wild type (white bars) are indicated. (G and H) Quantitation of methylation interference assays on the sense (G) and antisense (H) strands of PscpA. Nucleotides exhibiting a reduction in percentage bound versus free presented from 5′ to 3′. The values are averages of two independent experiments. The broken line in all panels denotes 75% of either wild-type binding or luciferase activity.