Abstract
Colicin M (Cma) is a protein toxin produced by Escherichia coli that kills sensitive E. coli cells by inhibiting murein biosynthesis in the periplasm. Recombinant plasmids carrying cbrA (formerly yidS) strongly increased resistance of cells to Cma, whereas deletion of cbrA increased Cma sensitivity. Transcription of cbrA is positively controlled by the two-component CreBC system. A ΔcreB mutant was highly Cma sensitive because little CbrA was synthesized. Treatment of CbrA-overproducing cells by osmotic shock failed to render cells Cma sensitive because the cells were resistant to osmotic shock. In a natural environment with a growth-limiting nutrient supply, cells producing CbrA defend themselves against colicin M synthesized by competing cells. Isolated CbrA is a protein with noncovalently bound flavin adenine dinucleotide. Sequence comparison and structure prediction assign the closest relative of CbrA with a known crystal structure as digeranylgeranyl-glycerophospholipid reductase of Thermoplasma acidophilum. CbrA is found in Escherichia coli, Citrobacter, and Salmonella bongori but not in other enterobacteria. The next homologs with the highest identity (over 50%) are found in the anaerobic Clostridium botulinum group 1 and a few other Firmicutes.
INTRODUCTION
Colicin M (Cma) is frequently encoded on plasmids of naturally occurring Escherichia coli isolates (11). The encoding gene, cma, usually forms an operon with the gene encoding colicin B, cba. Transcription of both cma and cba is regulated by the SOS response through an SOS box in front of cba (23, 32).
Sensitive E. coli cells take up Cma into the periplasm, where it inhibits murein biosynthesis by interfering with lipid carrier recycling (16, 37). Cma cleaves the phosphate ester bond between the precursor of murein biosynthesis and the lipid carrier, resulting in PP-MurNAc(pentapeptide)-GlcNAc, which is not incorporated into the growing murein layer, and undecaprenol, which no longer serves as a lipid carrier (13). Murein synthesis is therefore inhibited, and the cells lyse.
Cma is imported through the outer membrane transporter FhuA, which is coupled to the energy-providing electrochemical potential of the cytoplasmic membrane through the activity of the TonB, ExbB, and ExbD proteins (Ton system [6]). In the uptake of Cma, TonB interacts with both FhuA and Cma (33). Mutations in any of these proteins confer resistance to Cma. Other mutations also lead to Cma resistance, such as mutations in tolM (tolerance to colicin M [5, 38]). This gene was later identified as fkpA, which encodes a periplasmic chaperone/prolyl cis-trans isomerase (21). Point mutations in the isomerase domain of FkpA and fkpA deletion strains are completely resistant to Cma. FkpA is essential for Cma action and is required only for Cma and for no other tested colicin.
Cma has a compact structure, as revealed in the crystal structure (47), and must unfold to be translocated across the outer membrane. This change in Cma conformation most likely occurs upon binding to FhuA (21). In the periplasm, Cma interacts with FkpA, which likely is involved in the refolding of Cma, as indicated by the accelerated refolding of denatured Cma in vitro in the presence of FkpA (21). We previously proposed that unfolding of Cma during import involves a trans to cis isomerization of a proline bond, which is then cis to trans isomerized by FkpA during Cma refolding in the periplasm (20). This proposal is supported by the inactivity of Cma mutants in which other amino acids substitute for one specific proline residue (20) located outside the Cma active center (3, 20, 33) and by the high rate of cis-trans isomerization of this proline bond built into a synthetic peptide by FkpA (20).
Prior to the identification of tolM as fkpA, we had attempted to characterize tolM by cloning the mutated tolM gene on plasmids. Recombinant plasmids in different vectors that conferred high but not complete resistance to Cma were obtained, but the cloned genes were not identified. Here we report that the cloned gene in question is not tolM but rather yidS, now designated cbrA, and show that overproduction of CbrA renders cells resistant to Cma.
MATERIALS AND METHODS
Strains and plasmids.
The strains and plasmids used in this study are listed in Table 1. Plasmid pEG5005 contains a mini-Mu element and was used for in vivo cloning of tolM (fkpA) as described in reference 15. One of the recombinant plasmids obtained that conferred colicin M tolerance was pKH35 (with vector pACYC184), which is a subclone of pEG5005-derived plasmids.
Table 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| BW25113 | Δ(araD-araB)567 ΔlacZ4787(∷rrnB-3) λ− rph-1 Δ(rhaD-rhaB)568 hsdR514 | Keio Collection |
| JW5631 | Same as BW25113, but ΔcbrA | Keio Collection |
| JW4361 | Same as BW25113, but ΔcreB | Keio Collection |
| AB2847 | aroB tsx malT thi | University of Tübingen |
| BL21(DE3) | F− ompT gal dcm hsdB(rB− mB−) λ(DE3) lon lacI lacUV5-phage T7 gene 1 | 47 |
| Plasmids | ||
| pKH35 | cbrA on pACYC184 Cmr Tetr | This study |
| pSH150 | cbrA on pET-28(+) | This study |
| pHK569 | pHSG576 Cmr with a BamHI-PstI fragment of pGP1-2 | This institute |
| pET-28b(+) | T7 promoter lacI Kanr | Novagen |
| pES7 | cba cbi on pACYC184 | 39 |
| pKH55 | pcbrA-lacZ promoter fusion on pRS425, Ampr | This study |
| pRS425 | ′lacZ without promoter, Ampr | 41 |
| pAB115 | cbrA on pWSK29, Ampr | This study |
Plasmid pSH150 was constructed by introducing a 5′ NcoI and a 3′ XhoI cleavage site into pAB115 by PCR with Phusion high-fidelity polymerase (Finnzymes Oy, Vantaa, Finland) with the primers 5′-GAATGTGAGTGAAACCATGGAACA-TTTCGACGTG-3′ and 5′-CGATGTGAAGCGCGTCTCGAGATCCTTCAACTGTG-3′. The resulting DNA fragment comprising cbrA was cloned into pET28b cleaved with NcoI and XhoI. Plasmid pKH55 was constructed according to reference 48. The primers 5′-GGTACTGAATTCGAGTTCATGCATTACATGG-3′ and 5′-GCATCAGGATCCATCTTTTCACTCACATTCATCACG-3′ were used to amplify a 580-bp DNA fragment from chromosomal E. coli DNA. The fragment was digested with EcoRI and BamHI and ligated into the digested vector pRS415 so that the Met codon of cbrA became the start codon of lacZ. The expression of lacZ from plasmid pKH55 was very low. In plasmid pHK569, the kanamycin resistance marker of pGP1-2 (44) was replaced by a chloramphenicol resistance marker (H. Killmann, University of Tuebingen).
Growth of cells.
Cells were grown at 37°C aerobically or anaerobically in LB medium (10 g tryptone, 5 g yeast extract, and 5 g NaCl per liter) or in M9 medium (12.8 g Na2HPO4 · 7H2O, 3 g KH2PO4, 0.5 g NaCl, 1 g NH4Cl, 2 mM MgSO4, 0.1 mM CaCl2, 0.4% glucose, and 0.0002% thiamine per liter) supplemented, when required, with 50 μg/ml kanamycin, 40 μg/ml chloramphenicol, or 75 μg/ml ampicillin. Anaerobic growth conditions were created by filling 2.5-ml tubes with the medium, inoculating the medium, and tightly sealing the tubes. Growth was monitored by measuring the optical density at 578 nm (OD578).
To test the effect of Cma, Cba, or chemicals, overnight cultures grown aerobically or anaerobically were diluted and incubated with shaking at 200 rpm at 37°C to an OD578 of 0.3, at which Cma, Cba, or chemicals were added. Incubation was continued, and growth was monitored. OD578 was determined after 5 h of growth.
Colicin sensitivity assays.
Cma was overexpressed and purified as previously described (47). Cma sensitivity of cells was determined with purified Cma or with a crude cell extract in which Cma was by far the most prominent protein (19). Cba was overexpressed and its activity was determined as previously described (20). Crude extracts of Cma and Cba each contained approximately 1 mg colicin per ml. Strains were tested for colicin sensitivity either by spotting 10 μl of a 10-fold dilution series of the colicin onto agar plates seeded with the strain or by adding the colicin to liquid cultures (final concentration, 1 μg/ml). Purified Cma was transferred into the periplasm by osmotic shock as described previously (19, 20).
Sensitivity of cbrA-overproducing cells to colicins A, B, D, E1, E3, Ib, K, l, N, S4, U, 5, and 10 was determined by spotting 10 μl of a crude cell extract of each colicin-producing strain onto LB agar plates seeded with E. coli AB2847 carrying both plasmid pSH150, which carries cbrA, and plasmid pHK569, which encodes the T7 polymerase (43) under the control of the heat-sensitive λcI857 repressor (21). E. coli AB2847(pSH150)(pHK569) cells were grown at 30°C to an OD578 of 0.4, shifted to 42°C for 30 min, and then shifted to 37°C for 3 h. Cells were then spread on nutrient agar plates on which 10 μl of each colicin solution was dropped. The plates were incubated overnight at 37°C and inspected for lysis zones. E. coli AB2847 lacking plasmids was used as a control.
Determination of β-galactosidase activity.
For aerobically grown cells carrying pHK55 pcbrA-lacZ, cultures in M9 medium or in LB medium were incubated at 37°C to an OD578 of 0.6. Cells of 1 ml of culture were harvested by centrifugation at 4°C for 10 min, and the sediment was suspended in 1 ml buffer Z containing 10 μl 0.1% SDS and 20 μl chloroform (24). The suspensions were vortexed for 10 s and then incubated for 8 min at 28°C, after which 0.2 ml o-nitrophenyl-β-d-galactopyranoside (ONPG; 4 mg/ml in buffer Z) was added. After 150 min of incubation at 28°C, the reaction was stopped by adding 0.5 ml 1 M Na2CO3. The samples were centrifuged for 10 min at 4°C, and the A420 of the o-nitrophenol produced by cleavage of ONPG was measured.
For anaerobic growth, cells carrying pHK55 pcbrA-lacZ were incubated overnight in tightly sealed tube cultures in M9 medium or in LB medium to an OD578 of 0.6, and β-galactosidase activity was determined as described above.
Determination of undecaprenyl phosphate.
Cells of E. coli BW25113(pSH150)(pHK569) and E. coli JW5631 cbrA were grown in 1 liter of LB medium. Transcription of cbrA on pSH150 by the T7 polymerase encoded on pHK569 was induced by incubation for 30 min at 42°C. Cultivation was continued for 3 h at 37°C to an OD578 of 1.8. Cells from 100-ml cultures were harvested and stored at −20°C. The cell sediment was treated with 60% KOH in methanol-water for 1 h at 95°C to convert the isoprenoid phosphate esters into the monophosphate and the isoprenoids were extracted as described by Barreteau et al. (3).
Purification of CbrA.
E. coli BL21(DE3)(pSH150) carrying plasmid-borne cbrA was grown in 1.6 liters of LB medium to an OD578 of 0.5, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM, and the cells were further incubated for 3 h. Cells were harvested by centrifugation and suspended in 50 ml HSG buffer (20 mM HEPES, 200 mM NaCl, 10% glycerol) supplemented with 10 mM MgCl2, 1 mg DNase, and 1 tablet of complete serine/cysteine protease inhibitor cocktail, free of EDTA (Roche, Mannheim, Germany). Cells were disrupted in a French press, and the suspension was centrifuged for 45 min at 126,000 × g at 4°C. The sediment was suspended in HSG buffer supplemented with 1% undecyl maltoside (Glycon Biochemicals, Luckenwalde, Germany) and the protease inhibitor mixture and stirred overnight at 4°C. The suspension was centrifuged, and the CbrA-containing supernatant was applied to a 1-ml nickel-nitrilotriacetic acid (Ni-NTA) agarose column (GE Healthcare, Munich, Germany) equilibrated with HSG buffer containing 0.1% undecyl maltoside and 20 mM imidazole. The column was washed with equilibration buffer, and proteins were eluted with HSG buffer containing 0.1% undecyl maltoside and 200 mM imidazole, followed by HSG buffer containing 0.1% undecyl maltoside and 1 M imidazole. The eluted fractions were analyzed by SDS-PAGE as described previously (19).
Determination of FAD.
The supernatant of the Ni-NTA agarose fraction in HSG buffer containing 0.1% undecyl maltoside and 1 M imidazole was analyzed spectrophotometrically in a Perkin-Elmer Lambda 25 UV-visible (UV-Vis) spectrometer, and its spectrum was compared with that of commercial flavin adenine dinucleotide (FAD). In addition, the supernatant was examined by high-performance liquid chromatography (HPLC). Aliquots (5 μl) were injected onto an HPLC column (125-mm by 3-mm inside diameter [i.d.]; precolumn, 20-mm by 3-mm i.d.) packed with 5-μm Nucleosil100-C18 (Maisch, Ammerbuch, Germany) and separated with a linear gradient of 0.1% o-phosphoric acid in 4.5 to 100% acetonitrile in 15 min at a flow rate of 0.85 ml/min, followed by 3 min at 100% acetonitrile. HPLC was calibrated with commercial FAD under the same conditions as the supernatant.
In addition, FAD was determined by mass spectroscopy. The supernatants were examined with liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI-MS) using a Nucleosil 100-C18 column (3 μm; 100 by 2 mm) fitted with a guard column filled with Nucleosil 100-C18 (3 μm; 20 by 2 mm) coupled to an ESI mass spectrometer (LC/MSD Ultra Trap System XCT 6330; Agilent Technology). Analysis was performed at a flow rate of 0.4 ml min−1 with a linear gradient from 10% to 100% of solvent B in 15 min (solvent A, 0.1% formic acid in water; solvent B, 0.06% formic acid in acetonitrile). Detection was carried out at 230, 260, 280, 360, and 435 nm (±10 nm). Electron spray ionization (positive and negative ionization) in ultrascan mode with a capillary voltage of 3.5 kV and a heated temperature of 350°C was used. Commercial FAD was used for calibration. The HPLC-MS analysis was performed by Marcell Wagner and Andreas Kulik, IMIT, University of Tuebingen.
Bioinformatics.
Homologs of the CbrA protein of E. coli K-12 (GI 161784314) were identified by searching the nonredundant database at NCBI with PSI-BLAST (1). FAD-binding oxidoreductases with the Rossmann fold (35) were retrieved as the closest homologs. The 2,000 most similar sequences obtained by three iterations of PSI-BLAST were clustered in CLANS with BLAST as a comparison tool (14). Clustering was performed with default settings and a P value cutoff of 1.0e−15.
HHpred, a method for detecting remote homologs based on the comparison of profile hidden Markov models (42), was used to search the Protein Data Bank (PDB [4]) for homologs of known structure. A homology model of CbrA was generated with MODELLER (36) using the closest hit of the Protein Data Bank, relying on the alignment provided by HHpred. Molecular structures were rendered using PyMol (http://pymol.org).
RESULTS
Overexpressed cbrA confers resistance to Cma.
Earlier attempts to clone the mutated tolM (fkpA) gene from the E. coli genome resulted in plasmid pKH35, which conferred partial resistance of cells to Cma. Whereas a 105-fold-diluted Cma solution yielded a clear lysis zone on LB agar plates seeded with E. coli AB2847, a 103-fold dilution and lower dilutions of Cma yielded only turbid lysis zones on plates seeded with E. coli AB2847(pKH35) (Table 2). Strains carrying pKH35 were therefore more than 103-fold less sensitive to Cma than the strain not carrying the recombinant plasmid. pKH35 was derived from the pACYC184 vector with 18 copies per cell (10).
Table 2.
Cma sensitivities of E. coli strains transformed with plasmids carrying cbrA
| Strain | Diam of lysis zone (cm) with 10-fold dilution series |
||||||
|---|---|---|---|---|---|---|---|
| 100 | 101 | 102 | 103 | 104 | 105 | 106 | |
| AB2847 | 2 | 1.6 | 1.4 | 1.05 | 0.9 | 0.7 | 0.8a |
| AB2847(pKH35) | 1.7a | 1.5a | 1.1a | 0.9a | |||
| AB2847b | 1.9 | 1.6 | 1.4 | 1.2 | 0.8 | 0.7a | |
| AB2847(pSH150), uninduced | 1.7a | 1.3a | 1.0a | 0.9a | |||
| AB2847(pSH150), induced | 0 | 0 | 0 | 0 | |||
| BW25113 | ND | 1.7 | 1.5 | 1.3 | 1.0 | 0.9a | |
| JW5631 ΔcbrA | ND | 1.8 | 1.5 | 1.3 | 1.0 | 0.8a | |
Turbid lysis zone.
In this and the four following experiments, a less active Cma solution was used. An aliquot of a 10-fold dilution series of the purified Cma stock solution was spotted on an LB agar plate seeded with one of the strains to be tested. Cells carrying pSH150 also carried pHK569, which is a derivative of pGP1-2 (44) and encodes the T7 polymerase under the control of the temperature-sensitive λcI857 repressor. ND, not determined. The data were fully reproducible in several experiments.
The insert in pKH35 contained the open reading frame which was identical to the yidS (cbrA) gene of the E. coli genome. Overexpression of cbrA cloned downstream of the phage T7 gene 10 promoter on pSH150 in cells also producing the phage T7 RNA polymerase resulted in complete resistance to Cma (Table 2). These results suggested that Cma sensitivity is controlled by CbrA. In LB medium, the Cma sensitivity of the cbrA+ strain BW25113 was the same as that of the ΔcbrA mutant JW5631 (Table 2). As is shown below, cbrA is only very poorly transcribed in LB medium.
The level of Cma resistance depends on the level of CbrA expression.
The above-described results suggested that the level of Cma resistance is correlated with the level of CbrA synthesis. cbrA is preceded by the repeat sequence TTCACnnnnnnTTCAC (2), which serves as a binding site of the transcriptional regulator CreB (8). CreBC is a global two-component regulator of gene expression, with CreC as a sensor of unknown signals. CreBC-controlled transcription is initiated when cells are shifted from a rich medium to a poor medium, when cells are grown in minimal medium with fermentable glycolytic carbon sources, or when cells are grown aerobically with fermentation products as carbon sources (8, 27).
Since cbrA transcription should be induced when cells are under nutrient-limiting conditions, as usually occurs in natural environments, we tested the level of Cma resistance in rich LB medium (Table 3) and in glucose-M9 minimal medium (Fig. 1; Table 3) in the presence and absence of Cma. In M9 medium, E. coli BW25113 (i.e., cbrA+) was partially resistant to 1 μg/ml Cma, as indicated by the increase in optical density, which reached 0.58 after 150 min. E. coli BW25113(pKH35) (plasmid-borne cbrA) had a higher Cma resistance (OD578 = 0.91). In LB medium, E. coli BW25113 was fully sensitive to Cma (OD578 = 0.08) and E. coli BW25113(pKH35) was resistant (OD578 = 1.01). In contrast, the ΔcbrA mutant JW5631 was fully sensitive to Cma in both M9 and LB medium (OD578 = 0.07 for both). These results suggested that chromosomally encoded cbrA caused partial resistance to Cma in M9 medium but not in LB medium, that overexpression of plasmid-carried cbrA enhanced Cma resistance, and that lack of cbrA rendered cells Cma sensitive in both media. The level of CbrA that conferred Cma resistance depended on the medium, whereas overexpression by plasmid-borne cbrA was sufficiently high in both media to confer Cma resistance.
Table 3.
Comparison of CbrA-mediated Cma resistance in rich LB medium and minimal M9 mediuma
| Strain | Medium | OD578 after 150 min of growth |
|
|---|---|---|---|
| Aerobic | Anaerobic | ||
| BW25113 | LB | 2.70 | ND |
| BW25113 + Cma | LB | 0.08 | ND |
| JW5631 ΔcbrA | LB | 2.38 | ND |
| JW5631 ΔcbrA + Cma | LB | 0.07 | ND |
| BW25113(pKH35) | LB | 2.08 | ND |
| BW25113(pKH35) + Cma | LB | 1.01 | ND |
| BW25113 | M9 | 0.98 | 0.74 |
| BW25113 + Cma | M9 | 0.58 | 0.70 |
| JW4361 ΔcreB | M9 | 0.94 | 0.74 |
| JW4361 ΔcreB + Cma | M9 | 0.07 | 0.14 |
| JW5631 ΔcbrA | M9 | 1.05 | 0.77 |
| JW5631 ΔcbrA + Cma | M9 | 0.07 | 0.08 |
| BW25113(pKH35) | M9 | 0.96 | 0.64 |
| BW25113(pKH35) + Cma | M9 | 0.91 | 0.65 |
| JW4361 ΔcreB(pKH35) | M9 | 0.84 | 0.69 |
| JW4361 ΔcreB(pKH35) + Cma | M9 | 0.14 | 0.68 |
Cells were grown in rich LB medium or in glucose-M9 minimal medium to an OD578 of 0.3, when Cma (1 μg/ml) was added to the cultures as indicated. Growth was determined by measuring the OD578. Sensitivity to Cba (1 μg/ml) was tested in parallel; mutants and strain BW25113 cbrA+ had the same Cba sensitivity (data not listed). ND, not determined. The data varied ±5% in repeated experiments.
Fig 1.

Effect of Cma and Cba on the growth of E. coli cells encoding cbrA (cbrA+), lacking cbrA (ΔcbrA), and overexpressing cbrA (on pKH35). Growth of the indicated strains in M9 minimal medium in the absence and presence of 1 μg/ml Cma (+ Cma) or Cba (+ Cba) was monitored spectrophotometrically. Cma or Cba was added to the indicated exponentially growing cultures at an OD578 of 0.3 (arrow). The curves are representative of several repeated experiments.
cbrA transcription is under the control of CreB (2, 7). If the level of CbrA controls Cma sensitivity, a ΔcreB mutant should reduce cbrA transcription and render cells Cma sensitive. This was indeed the case. The ΔcreB mutant JW4361 was fully sensitive to Cma in M9 medium (OD578 = 0.08 [Table 3]). E. coli JW4361(pKH35) (i.e., ΔcreB and plasmid-borne cbrA) had regained Cma resistance (OD578 = 0.68). CreBC-controlled genes are believed to be more strongly expressed during anaerobic growth than during aerobic growth (2). Apart from the lower growth rate under anaerobic conditions, the values obtained for aerobic and anaerobic cultures are similar, with one exception. E. coli JW4361(pKH35) was more resistant to Cma when grown anaerobically than when grown aerobically (OD578 = 0.14 versus 0.68 [Table 3]). During anaerobic growth, cbrA on pKH35 was sufficiently expressed to confer Cma resistance even though the CreB transcription initiator was lacking.
Colicin B (Cba) was used as a control to test whether the different growth rates in LB and M9 media affected the degree of colicin sensitivity and to see whether CbrA affected sensitivity to colicins other than Cma. Wild-type and mutant cells treated with Cba stopped growth to the same extent regardless of whether they were grown in LB or M9 medium (Fig. 1 shows growth in M9 medium). Note that the optical density did not decrease in these cases, because Cba is a pore-forming colicin that does not immediately cause cell lysis (34).
To further test the specificity of cbrA-related Cma resistance, E. coli AB2847(pSH150)(pKH569) was spread on LB agar plates onto which crude extracts of various colicins were added. Transcription of cbrA on pSH150 was under the control of the gene 10 promoter of phage T7, which was transcribed by the T7 RNA polymerase encoded on pKH569. Transcription of the RNA polymerase gene was induced at 42°C prior to the addition of the colicins. Colicins A, B, D, E1, E3, Ib, K, L, N, S4, U, 5, and 10 each formed clear lysis zones (data not shown), in contrast to Cma, which formed no lysis zone. These results indicate that Cma resistance caused by overexpressed CbrA is Cma specific.
Regulation of cbrA transcription.
To further correlate the level of cbrA expression with Cma resistance, we fused the upstream region of cbrA including the repeat sequence TTCACAAGGACTTCAC with the E. coli lacZ gene. Cells were transformed with the resulting plasmid, pKH55 carrying pcbrA-lacZ, and the levels of β-galactosidase activity in cells growing aerobically or anaerobically in LB medium and M9 minimal medium were measured (Table 4). The β-galactosidase values of LB-grown strains were all much lower than the values of M9-grown cells. For example, the level of β-galactosidase activity in cells of E. coli BW25113(pKH55) grown in M9 medium was 8-fold higher than that in cells grown in LB medium. The lack of creB reduced β-galactosidase activity 4.5-fold in cells growing aerobically and 9.1-fold in cells growing anaerobically. The values of aerobically grown cells were similar to the values of anaerobically grown cells.
Table 4.
β-Galactosidase activities of E. coli strains carrying pcbrA-lacZ fusions on plasmid pKH55a
| Strain | Medium | Growth | Activity (U) |
|---|---|---|---|
| BW25113(pKH55) | M9 | Aerobic | 16.2 |
| JW4361 ΔcreB(pKH55) | M9 | Aerobic | 3.6 |
| JW5631 ΔcbrA(pKH55) | M9 | Aerobic | 14.4 |
| BW25113(pKH55) | M9 | Anaerobic | 14.5 |
| JW4361 ΔcreB(pKH55) | M9 | Anaerobic | 1.6 |
| JW5631 ΔcbrA(pKH55) | M9 | Anaerobic | 13.3 |
| BW25113(pKH55) | LB | Aerobic | 2.1 |
| JW4361 ΔcreB(pKH55) | LB | Aerobic | 0.7 |
| JW5631 ΔcbrA(pKH55) | LB | Aerobic | 1.5 |
Plasmid pKH55 encodes the pcbrA-lacZ operon fusion under the control of CreB. The values varied ±5% in repeated experiments.
To avoid plasmid copy number effects, λpcbrA-lacZ lysogens were examined. Colonies of cells carrying this construct were only slightly blue on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) plates, and the β-galactosidase levels were much lower than those of cells carrying plasmid-encoded pcbrA-lacZ (data not shown). A proteome analysis of E. coli grown in rich medium did not reveal CbrA (Boris Macek, Proteome Center, University of Tuebingen, personal communication), which further indicates the very low expression level of chromosomally encoded CbrA.
CbrA alters the E. coli cell envelope.
To examine whether overexpressed CbrA inhibits Cma uptake, the Cma transport system via FhuA, TonB, ExbB, and ExbD was bypassed by osmotic shock treatment of cells. This widely used procedure to shock proteins out of the periplasm (31) can also be used to shock proteins into the periplasm and has been successfully used to shock Cma into the periplasm of Cma import mutants (5, 19). As expected, E. coli BW25113 and JW5631 osmotically shocked in the presence of Cma were highly sensitive to Cma (less than 0.01% survival [Table 5]). E. coli BW25113(pKH35) (i.e., cbrA on pACYC184) osmotically shocked in the presence of Cma showed a rather high level of resistance to Cma (34% survival). The resistance to Cma was caused by the resistance of the cells to osmotic shock, as shown by the survival of 54% of cells of this strain osmotically shocked in the absence of Cma, compared to 18% survival of E. coli BW25113 cells lacking plasmid pKH35. E. coli JW5631, which differs from strain BW25113 only by its lack of chromosomally encoded cbrA, incubated with Cma and without osmotic shock was even more sensitive to Cma than strain BW25113, having 18 times fewer survivors. In contrast, in cells incubated with Cma and without osmotic shock, overproduction of plasmid-encoded cbrA in strain BW25113(pKH35) increased the resistance to Cma 76.7-fold compared to that of cbrA+ cells and 1,380-fold compared to that of the ΔcbrA mutant. The shock resistance of strain BW25113(pKH35) suggests that CbrA alters the cell envelope, possibly the outer membrane, such that Cma can be only poorly transferred into the periplasm by osmotic shock. Transformation of E. coli BW25113 by the vector pACYC184 did not confer shock resistance (Table 5).
Table 5.
Cell survival after osmotic shock in the presence and absence of Cmaa
| Strain | % of cells surviving osmotic shock |
% of cells surviving incubation with Cma without osmotic shock | |
|---|---|---|---|
| With Cma | No Cma | ||
| BW25113 | 0 | 18 | 0.9 |
| JW5631 ΔcbrA | 0 | 17 | 0.05 |
| BW25113(pKH35) | 34 | 54 | 69 |
| BW25113(pACYC184) | 0 | 5 | 1.3 |
Cells were shifted from a medium of high osmolarity to a medium of low osmolarity (osmotic shock) in the presence or absence of Cma (10 μg/0.34 ml). Cells were plated on LB agar, and the colonies formed by the surviving cells were counted. The values are relative to the values of untreated cultures (100% survival). Values of <0.01% are listed as 0%. They varied ±10% in repeated experiments.
Sensitivity of the ΔcbrA mutant to chemicals.
In a phenotype microarray analysis of E. coli mutants with deletions in all two-component systems, mutants with changes in cbrA displayed a greater resistance to hydroxylamine and hypersensitivity to ofloxacin, which suggests that membrane permeability is affected (48). Therefore, we compared the anaerobic growth in M9 medium containing 1 μg/ml ofloxacin of E. coli strains BW25113, JW6531 ΔcbrA, JW4361 ΔcreB, and BW25113(pKH35) (plasmid-carried cbrA). The cbrA and creB deletion strains grew as well as strain BW25113, but the growth of strain BW25113(pKH35) was slightly reduced, by 24%. Hydroxylamine (50 μg/ml) inhibited the anaerobic growth in M9 minimal medium of all the strains tested to the same extent (36 to 39%). These data differ from the published results, but the experimental conditions we used (growth rates) were not identical to the conditions formerly used (respiration rates) (48). Moreover, the former study found phenotypes not for creABCD mutants but only for the cbrA mutant, which led to the hypothesis that additional controls need further studies.
We also tested the effect of compounds known to inhibit E. coli mutants with defects in the outer membrane, e.g., tol mutants. Growth of E. coli BW25113, BW25113(pKH35), JW5631 ΔcbrA, and JW4361 ΔcreB in M9 minimal medium containing 0.1 μg/ml polymyxin was reduced to similar levels, i.e., the OD578 ranged from 0.12 to 0.14, compared to the OD578 range of 0.53 to 0.61 of the strains grown without polymyxin. Incubation of the cells in M9 medium containing 1 mM EDTA or 1% sodium cholate only slightly reduced the growth rate of all the strains, to similar extents (data not shown). In addition, strains JW5631 ΔcbrA and BW25113 were similarly sensitive to albomycin (data not shown), which, like Cma, is actively taken up by E. coli cells via the FhuA and Ton system (18). These results indicate that a lack or overproduction of CbrA caused no general defect in outer membrane permeability.
Purification of the CbrA protein.
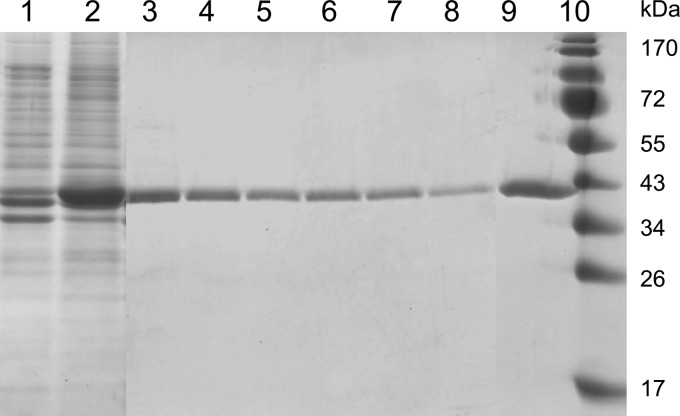
To identify the cbrA gene product, E. coli BL21(DE3) was used to overexpress cbrA cloned downstream of the phage T7 promoter on plasmid pET-28b(+) by the IPTG-inducible chromosomally encoded T7 RNA polymerase. The synthesized protein carried a C-terminal His6 tag. Isolation of the CbrA protein was difficult because CbrA formed inclusion bodies that only partially dissolved in various detergents, 0.1 M Na2CO3, 6 M guanidinium chloride, or 8 M urea. CbrA was found only in the sediment of disrupted cells after centrifugation (Fig. 2). Only 1% undecyl maltoside resulted in a soluble fraction of CbrA-His6; the protein adsorbed to an Ni-NTA column and was eluted in two steps with HSG buffer and 0.1% undecyl maltoside containing 0.2 and 1 M imidazole (Fig. 2). The position of the protein band on the SDS-polyacrylamide gel agreed with the predicted molecular mass of 40 kDa. The eluted protein formed a precipitate that was yellow in fractions containing 0.2 M imidazole and white with a yellow supernatant in fractions containing 1 M imidazole. HPLC of the supernatant resulted in a major peak at the position of authentic commercial FAD (Fig. 3A). Coupling of HPLC with mass spectrometry revealed the theoretical mass of 785 Da for the oxidized form of FAD. Also, the UV-Vis absorption spectrum of the supernatant was typical for oxidized FAD (Fig. 3B). These results indicate that CbrA contains noncovalently bound FAD. We assume that imidazole weakly binds to the FAD binding site of CbrA and that high concentrations of imidazole are required to dissociate FAD from CbrA. It remains bound to precipitated CbrA because CbrA aggregation does not necessarily imply denaturation. The predicted CbrA structure contains a surface-exposed hydrophobic region (see Fig. 5) through which it may aggregate.
Fig 2.
SDS-PAGE analysis of the CbrA purification steps. The cbrA gene cloned in vector pET-28b(+) in E. coli BL21(DE3) was overexpressed after induction with IPTG; synthesized CbrA carried a C-terminal His6 tag. Shown are the sediments of disrupted and centrifuged uninduced (lane 1) and IPTG-induced (lane 2) cells, fractions obtained after Ni-NTA agarose column chromatography in HSG buffer and 0.1% undecyl maltoside containing 0.2 M imidazole (lanes 3 to 8) and 1 M imidazole (lane 9), and molecular size markers (lane 10).
Fig 3.
Determination of FAD in the purified Cba protein. (A) HPLC of the supernatant of the precipitated fraction after Ni-NTA agarose column chromatography in HSG buffer, 0.1% undecyl maltoside, and 1 M imidazole recorded at 260 nm. 1, FAD; 2, riboflavin. (B) UV-Vis spectrum of the supernatant fraction.
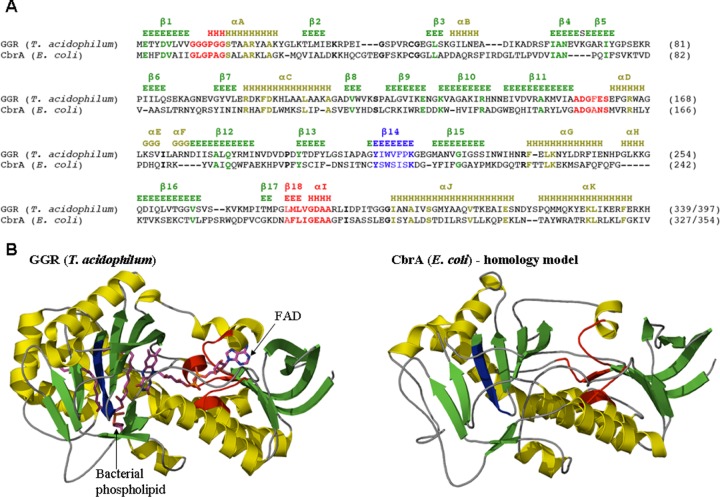
Fig 5.
Conservation of functional residues suggests an isoprenoid substrate for CbrA. (A) The sequence alignment of E. coli CbrA and the closest homolog of known structure, geranylgeranyl reductase (GGR) from Thermoplasma acidophilum (PDB identifier 3OZ2 [46]), emphasizes the conservation of residues associated with substrate binding. Common motifs involved in nucleotide binding are shown in red. Identical residues in helices and strands are shown in yellow and green, respectively, and are annotated according to reference 47. The conserved lipid binding site is colored in blue. The alignment was obtained from HHpred (42) with a P value of 1.0e−48 and a pairwise identity of 20%. (B) The homology model of CbrA (right) shows a Rossmann fold (35) FAD-dependent oxidoreductase. GGR from T. acidophilum (left), which was used as a template, illustrates binding of an FAD moiety and of a bacterial lipid (48). The coloring is according to the sequence alignment shown in panel A.
Homology of CbrA and geranylgeranyl reductases (GGRs).
cbrA occurs rarely in bacterial genomes and shows an unusual distribution (see Fig. S1 in the supplemental material). It is found only in a few enterobacteria like E. coli, Citrobacter, and Salmonella bongori and not in other Salmonella strains or in other enterobacteria. The next cbrA homologs are found in the anaerobe Firmicutes Sebaldella termitidis and Elusimicrobium minutum, which have been isolated from termite intestine, and in Clostridium perfringens strains, Clostridium botulinum group I strains, and Clostridium sporogenes strains (51 to 55% sequence identity). The GC content of cbrA in C. botulinum deviates strongly from the low GC content of the genome, which may indicate a more recent lateral gene transfer. However, in the related C. perfringens, there is no difference in GC content observed. Sequences with identities below 45% are found in Helicobacter species. Some geranylgeranyl hydrogenases show 28% identity and lower. It is interesting to note that close relatives to cbrA are found in anaerobic and strictly anaerobic bacteria of Gram-negative and Gram-positive species, but in E. coli, expression of cbrA is not enhanced at anoxic conditions.
To further examine the evolutionary relationships of CbrA proteins and their homologs, we collected the 2,000 most similar proteins using PSI-BLAST and clustered the sequences with CLANS (1, 14). The cluster map shows that CbrA proteins belong to a large superfamily of FAD-binding proteins (Fig. 4), exemplified by p-hydroxybenzoate hydroxylase and many other FAD-dependent oxidoreductases with diverse functions (25). Among these, the closest homologs of CbrA are the GGRs, which are widely distributed in plants (22), archaea (28), and bacteria, especially proteobacteria, actinobacteria, and cyanobacteria (40). In contrast, the presence of CbrA orthologs is restricted mainly to E. coli strains and a few other proteobacteria, e.g., Shigella. However, the shared common ancestry of CbrA with FAD-dependent enzymes is consistent with its ability to bind FAD.
Fig 4.
Cluster map of the relationships of CbrA protein to its nearest homologs among FAD-binding oxidoreductases. Pairwise similarities of all sequences were computed with BLAST (1). The cluster map was constructed using the program CLANS (14). The clustering procedure uses all P values better than 1.0e−15. Darker lines between two dots, each of which represents one sequence, indicate lower BLAST P values. The map contains 2,000 sequences.
Using the CbrA sequence, we searched the Protein Data Bank (PDB [4]) for the closest homolog of known structure. The top hit of searches available on 4 February 2012 that clustered at a maximum of 70% pairwise sequence identity for proteins similar to CbrA was the GGR of the archaeon Thermoplasma acidophilum (PDB identifier 3OZ2) (46). HHpred retrieved a P value of 1.0e−49 and 20% pairwise sequence identity of CbrA and 3OZ2 using two iterations of PSI-BLAST for multiple-sequence alignment generation and activating the realignment with the MAC option. Additional searches with CbrA against the SCOP database (25), version 1.75, clustered at a maximum of 70% pairwise sequence identity confirmed the assignment of CbrA to the Rossmann fold type of FAD-binding oxidoreductases (12).
The crystal structure of the T. acidophilum GGR has been solved in complex with FAD and an endogenous bacterial phospholipid, which allowed identification of residues involved in substrate binding (46). Detailed analysis of the sequence similarity between CbrA and this GGR revealed conservation of residues crucial for binding not only of the FAD moiety but also of the lipid (Fig. 5A). This includes the conserved YxWxFP motif that aligns the double bond of the geranyl group with respect to FAD and is therefore important for substrate specificity. The conservation of this and other sequence motifs between CbrA and GGRs suggests that CbrA in E. coli may function similarly to GGR during the synthesis of archaeal lipids, namely, to saturate isoprenoid molecules (28, 29). Furthermore, the homology of CbrA, GGRs, and other FAD-dependent oxidoreductases predicts that CbrA also adopts a divergent Rossmann fold (12, 35) (Fig. 5B).
Lack of evidence that CbrA modifies undecaprenyl derivatives.
Similarity of CbrA with geranylgeranyl reductases suggested that CbrA might reduce undecaprenyl derivatives, including murein precursors which no longer may serve as a substrate for Cma, thus explaining CbrA-mediated Cma resistance. To test this hypothesis, undecaprenyl derivatives were extracted from cells and converted to undecaprenyl phosphate by treatment with KOH. HPLC analysis revealed only unmodified undecaprenyl phosphate in cells that overexpressed CbrA and cells that were lacking CbrA. These results do not support the notion that CbrA reduces double bonds of undecaprenyl-linked murein precursors, leading to lipid I and lipid II derivatives that are not cleaved by Cma.
DISCUSSION
Cma kills sensitive E. coli cells by inhibiting murein biosynthesis (13, 16, 37), which causes cell lysis. It also inhibits O-antigen biosynthesis (17), which, although harmless under laboratory conditions, is apparently detrimental under natural conditions since lipopolysaccharide rough strains of E. coli natural isolates are usually not found.
E. coli cells growing in the neighborhood of Cma-producing cells could protect themselves by inactivating the Cma uptake system. But loss of FhuA, TonB, ExbB, and ExbD or FkpA probably reduces the fitness of such mutants to an intolerable degree, since these functions were developed and preserved during evolution. In a screen for Cma-resistant mutants, target site mutants have not been found (38), which suggests that an altered Cma substrate does not serve as a precursor of murein biosynthesis.
Our study describes a novel means of protection against Cma—the CbrA protein. E. coli cells producing wild-type amounts of CbrA or overproducing the protein have a higher survival rate in the presence of Cma than cells lacking CbrA (18-fold or 1,380-fold higher survival, respectively [Table 5], no osmotic shock, with Cma). Depending on the level of overproduction, the amount of Cma required to form clear lysis zones differed 1,000-fold, and high CbrA production (pSH150) resulted in complete resistance (Table 2). Although transcription of chromosomal cbrA was very low even in M9 medium (data not shown), the amount of CbrA conferred partial Cma resistance (Table 3 and Fig. 1, BW25113 + Cma). This level of expression may be sufficient for cells to compete with Cma-producing clones in natural environments.
The cbrA promoter contains a CreBC-responsive sequence. Indeed, pcbrA-lacZ transcription was upregulated in LB and M9 media by creB, as revealed by the creB mutant with low expression (Table 4), and was correlated with the degree of Cma sensitivity (Table 3). CreBC-dependent upregulation of cbrA expression has been observed in reverse transcription-PCR (RT-PCR) and DNA microarray analyses, and our data supplement these earlier observations (2, 7).
Recently, another CreBC-regulated protein, CbrC (YieJ), that increases resistance to colicin E2 has been identified (7). A 95-fold overexpression of cbrC in a mutant carrying a point mutation in the CreC histidine kinase domain leads to survival at a colicin E2 concentration 16-fold higher than the concentration survived by the CreC parent strain. The cbrC gene under the control of the arabinose promoter confers an 8-fold-higher E2 resistance when transcription is induced by arabinose.
cbrA and cbrC do not form an operon. They are separated by 20 open reading frames on the E. coli genome. Their cbr designation is not based on a functional context but rather is derived from the common regulation by creBC. In fact, no function had been previously experimentally assigned to any of the nine genes shown to be controlled by CreBC (7). Our results, namely, the tolerance of CbrA-producing cells to Cma and the resistance to osmotic shock, suggest a function of CbrA in outer membrane structure. Similarly, an alteration in the membrane structure also applies to the colicin E2 tolerance conferred by overproduction of CbrC (7). Additional direct effects of CbrA on the target site of Cma cannot be excluded. However, in the case of colicin E2, direct effects at the target site are highly unlikely because the colicin acts as a DNase in the cytoplasm (9).
In contrast to CbrA and CbrC, which confer resistance to specific colicins, Tol proteins confer sensitivity to the entire group A colicins (26, 30). tol mutants show pleiotropic phenotypes, such as hypersensitivity to bile, detergents, toxic compounds, and antibiotics and resistance to filamentous phages. In contrast, the cbrA mutant was not sensitive to the chemicals tested, namely, ofloxacin, EDTA, cholate, and polymyxin. The only clear phenotype observed with respect to cell envelope permeability that was caused by CbrA was the increased resistance to osmotic shock when cbrA was overexpressed. Overproduction of CbrA did not affect the activity of the Ton system, required for Cma uptake, as both strain BW25113(pKH35) (plasmid-borne cbrA) and strain JW5631 ΔcbrA were sensitive to all tested TonB-dependent colicins and albomycin. cbrA deletion also did not alter sensitivity to Tol-dependent group A colicins.
Purification of the CbrA protein was difficult because under all tested conditions, CbrA formed inclusion bodies and precipitated after the solubilizing agents were removed. At high ionic conditions (1 M imidazole), the isolated yellow protein precipitate turned white and the supernatant became yellow, indicating the release of FAD from CbrA, as determined by UV-Vis spectroscopy, mass spectroscopy, and HPLC. Protein sequence comparisons predicted that CbrA is an FAD/NAD(P)-dependent oxidoreductase with polyisoprenoids as possible substrates. However, we found no suitable conditions for determining the enzymatic activity of the partially solubilized protein. Therefore, we could not determine whether the substrate of Cma—the C55 polyisoprenyl-PP-MurNAc(pentapeptide)-GlcNAc—is altered by CbrA. The polyisoprenoid resides in the cytoplasmic membrane. Although CbrA lacks predicted transmembrane regions, the insertion of a portion of CbrA into the cytoplasmic membrane cannot be excluded. Indeed, the strong aggregation of CbrA suggests a hydrophobic region at the protein surface through which it could contact the cytoplasmic membrane. A surface-exposed lipid binding region which could form the interface for aggregation and membrane association is predicted in CbrA (Fig. 5).
The closest relative of CbrA with a known crystal structure is the GGR from T. acidophilum (48). GGR converts unsaturated 2,3-di-O-geranylgeranylglyceryl phosphate to saturated 2,3-di-O-phytanylglyceryl phosphate. It is conceivable that CbrA catalyzes a similar reaction in reducing undecaprenyl phosphate or undecaprenyl-PP-MurNAc(pentapeptide)-GlcNAc (lipid II), the substrate of Cma. Hydrogenation could convert lipid II to a derivative that is no longer cleaved by Cma. Such a reaction would imply that the reduced form still functions as a substrate for murein biosynthesis, which involves translocation of the precursor across the outer membrane and insertion into the murein layer. However, analysis of undecaprenyl phosphate in a CbrA-overexpressing strain and a strain lacking CbrA revealed only unchanged undecaprenyl phosphate. Hydrogenation of lipids I and II into derivatives that are not cleaved by Cma does not seem to be the mechanism of CbrA-mediated Cma resistance. It is not excluded that CbrA catalyzes cis-trans isomerization of double bonds in undecaprenylphosphate which our assay would not have revealed. However, it is difficult to conceive that such an alteration of the murein precursors would not affect murein biosynthesis. Alternatively, reduction of polyisoprenoids could be a side reaction that leads to saturated compounds, as is the case for the saturated archaeal tetraetherlipoglycans that stabilize membranes against high temperature, extreme pH, and osmotic stress (45).
CbrA is not an essential protein for E. coli. cbrA mutants show no phenotype under laboratory conditions. The scarce occurrence of CbrA orthologs in genomes of other bacteria and the lack of flanking genes that could be related to CbrA function prevented identification of a likely function that could be tested. The identification of CbrA as a protein that confers a high resistance to Cma and renders cells resistant to osmotic shock assigned a membrane function to CbrA. This function became apparent when cells were grown under nutrient-limiting conditions—conditions usually found in nature. Such conditions are necessary for the activation of the CreBC two-component regulatory system, which stimulates cbrA transcription. Cma sensitivity may be a convenient tool to study CreBC-mediated gene transcription and, in particular, to identify the signal that regulates CreC phosphorylation and, in turn, CreB phosphorylation and transcription initiation.
Nearly half of the E. coli natural isolates produce colicins, and pColBM plasmids are among the most frequently occurring plasmids (11). The large pColBM plasmids encode virulence factors, such as iron transport systems, hemolysins, hemagglutinins, and a complement resistance factor, which are lost when pColBM is lost. pColBM is maintained by the colicin M immunity protein encoded on pColBM. The immunity protein protects the producer from being killed by its own Cma and Cma produced by surrounding cells. Cells lacking pColBM would be able to survive and compete with Cma-producing cells in natural environments by synthesis of CbrA.
Supplementary Material
ACKNOWLEDGMENTS
We thank Andrei Lupas for generously hosting the Max-Planck group, Marcell Wagner and Andreas Kulik for HPLC-MS analysis, Helga Wolff for technical assistance, and Karen A. Brune for critically reading the manuscript.
This work was supported by the Max Planck Society and the German Research Foundation (BR330/25-1).
Footnotes
Published ahead of print 6 July 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Avison MB, Horton RE, Walsh TR, Bennett PM. 2001. Escherichia coli CreBC is a global regulator of gene expression that responds to growth in minimal medium. J. Biol. Chem. 276:26955–26961 [DOI] [PubMed] [Google Scholar]
- 3. Barreteau H, et al. 2010. Deciphering the catalytic domain of colicin M, a peptidoglycan lipid II-degrading enzyme. J. Biol. Chem. 285:12378–12389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Berman HM, et al. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Braun V, Frenz J, Hantke K, Schaller K. 1980. Penetration of colicin M into cells of Escherichia coli. J. Bacteriol. 142:162–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Braun V, Patzer SI, Hantke K. 2002. Ton-dependent colicins and microcins: modular design and evolution. Biochimie 84:365–380 [DOI] [PubMed] [Google Scholar]
- 7. Cariss SJL, et al. 2010. YieJ (CbrC) mediates CreBC-dependent colicin E2 tolerance in Escherichia coli. J. Bacteriol. 192:3329–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cariss SJL, Taylor AE, Avison MB. 2008. Defining the growth conditions and promoter-proximal DNA sequences required for activation of gene expression by CreBC in Escherichia coli. J. Bacteriol. 190:3930–3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cascales E, et al. 2007. Colicin biology. Microbiol. Mol. Biol. Rev. 71:158–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chang AC, Cohen SN. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the p15A cryptic plasmid. J. Bacteriol. 134:1141–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Christenson JK, Gordon DM. 2009. Evolution of colicin BM plasmids: the loss of the colicin B activity gene. Microbiology 155:1645–1655 [DOI] [PubMed] [Google Scholar]
- 12. Dym O, Eisenberg D. 2001. Sequence-structure analysis of FAD-containing proteins. Protein Sci. 10:1712–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. El Ghachi MA, et al. 2006. Colicin M exerts its bacteriolytic effect via enzymatic degradation of undecaprenyl phosphate-linked peptidoglycan precursors. J. Biol. Chem. 281:22761–22772 [DOI] [PubMed] [Google Scholar]
- 14. Frickey T, Lupas AN. 2004. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics 20:3702–3704 [DOI] [PubMed] [Google Scholar]
- 15. Groisman EA, Casadaban MJ. 1986. Mini-mu bacteriophage with plasmid replicons for in vivo cloning and lac gene fusing. J. Bacteriol. 168:357–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harkness RE, Braun V. 1989. Colicin M inhibits peptidoglycan biosynthesis by interfering with lipid carrier recycling. J. Biol. Chem. 264:6177–6182 [PubMed] [Google Scholar]
- 17. Harkness RE, Braun V. 1989. Inhibition of lipopolysaccharide O-antigen synthesis by colicin M. J. Biol. Chem. 264:14716–14722 [PubMed] [Google Scholar]
- 18. Hartmann A, Fiedler HP, Braun V. 1979. Uptake and conversion of the antibiotic albomycin by Escherichia coli K-12. Eur. J. Biochem. 99:517–524 [DOI] [PubMed] [Google Scholar]
- 19. Helbig S, Braun V. 2011. Mapping functional domains of colicin M. J. Bacteriol. 193:815–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Helbig S, Patzer SI, Schiene-Fischer C, Zeth K, Braun V. 2011. Activation of colicin M by the FkpA prolyl cis-trans isomerase/chaperone. J. Biol. Chem. 286:6280–6290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hullmann J, Patzer SI, Römer C, Hantke K, Braun V. 2008. Periplasmic chaperone FkpA is essential for imported colicin M toxicity. Mol. Microbiol. 69:926–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Keller Y, Bouvier F, d'Harlingue A, Camara B. 1998. Metabolic compartmentation of plastid prenyllipid biosynthesis—evidence for the involvement of a multifunctional geranylgeranyl reductase. Eur. J. Biochem. 251:413–417 [DOI] [PubMed] [Google Scholar]
- 23. Köck J, Ölschläger T, Kamp RM, Braun V. 1987. Primary structure of colicin M, an inhibitor of murein biosynthesis. J. Bacteriol. 169:3358–3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Miller JH. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 25. Murzin AG, Brenner SE, Hubbard T, Chothia C. 1995. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247:536–540 [DOI] [PubMed] [Google Scholar]
- 26. Nagel de Zwaig R, Luria SE. 1967. Genetics and physiology of colicin-tolerant mutants of Escherichia coli. J. Bacteriol. 94:1112–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nikel PI, Zhu J, San KY, Méndez BS, Bennett GN. 2009. Metabolic flux analysis of Escherichia coli creB and arcA mutants reveals shared control of carbon catabolism under microaerobic growth conditions. J. Bacteriol. 191:5538–5548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nishimura Y, Eguchi T. 2006. Biosynthesis of archaeal membrane lipids: digeranylgeranylglycerophospholipid reductase of the thermoacidophilic archaeon Thermoplasma acidophilum. J. Biochem. 139:1073–1081 [DOI] [PubMed] [Google Scholar]
- 29. Nishimura Y, Eguchi T. 2007. Stereochemistry of reduction in digeranylgeranylglycerophospholipid reductase involved in the biosynthesis of archaeal membrane lipids from Thermoplasma acidophilum. Bioorg. Chem. 35:276–283 [DOI] [PubMed] [Google Scholar]
- 30. Nomura M, Witten C. 1967. Interaction of colicins with bacterial cells. III. Colicin-tolerant mutations in Escherichia coli. J. Bacteriol. 94:1093–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nossal NG, Heppel LA. 1966. The release of enzymes by osmotic shock from Escherichia coli in exponential phase. J. Biol. Chem. 241:3055–3062 [PubMed] [Google Scholar]
- 32. Olschläger T, Schramm E, Braun V. 1984. Cloning and expression of the activity and immunity genes of colicins B and M on pColBM plasmids. Mol. Gen. Genet. 196:482–487 [DOI] [PubMed] [Google Scholar]
- 33. Pilsl H, et al. 1993. Domains of colicin M involved in uptake and activity. Mol. Gen. Genet. 240:103–112 [DOI] [PubMed] [Google Scholar]
- 34. Pressler U, Braun V, Wittmann-Liebold B, Benz R. 1986. Structural and functional properties of colicin B. J. Biol. Chem. 261:2654–2659 [PubMed] [Google Scholar]
- 35. Rossmann MG, Moras D, Olsen KW. 1974. Chemical and biological evolution of nucleotide-binding protein. Nature 250:194–199 [DOI] [PubMed] [Google Scholar]
- 36. Sali A, Blundell TL. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779–815 [DOI] [PubMed] [Google Scholar]
- 37. Schaller K, Höltje JV, Braun V. 1982. Colicin M is an inhibitor of murein biosynthesis. J. Bacteriol. 152:994–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schaller K, Krauel A, Braun V. 1981. Temperature-sensitive, colicin M-tolerant mutant of Escherichia coli. J. Bacteriol. 147:135–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schramm E, Mende J, Braun V, Kamp RM. 1987. Nucleotide sequence of the colicin B activity gene cba: consensus pentapeptide among TonB-dependent colicins and receptors. J. Bacteriol. 169:3350–3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shpilyov AV, Zinchenko VV, Shestakov SV, Grimm B, Lokstein H. 2005. Inactivation of the geranylgeranyl reductase (ChlP) gene in the cyanobacterium Synechocystis sp. PCC 6803. Biochim. Biophys. Acta 1706:195–203 [DOI] [PubMed] [Google Scholar]
- 41. Simons RW, Houman F, Kleckner N. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85–96 [DOI] [PubMed] [Google Scholar]
- 42. Söding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113–130 [DOI] [PubMed] [Google Scholar]
- 44. Tabor S, Richardson CC. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. U. S. A. 82:1074–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. van de Vossenberg JL, Driessen AJ, Konings WN. 1998. The essence of being extremophilic: the role of the unique archaeal membrane lipids. Extremophiles 2:163–170 [DOI] [PubMed] [Google Scholar]
- 46. Xu Q, et al. 2010. Insights into substrate specificity of geranylgeranyl reductases revealed by the structure of digeranylgeranylglycerophospholipid reductase, an essential enzyme in the biosynthesis of archaeal membrane lipids. J. Mol. Biol. 404:403–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zeth K, Römer C, Patzer SI, Braun V. 2008. Crystal structure of colicin M, a novel phosphatase specifically imported by Escherichia coli. J. Biol. Chem. 283:25324–25331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhou L, Lei XH, Bochner BR, Wanner BL. 2003. Phenotype MicroArray analysis of Escherichia coli K-12 mutants with deletions of all two-component systems. J. Bacteriol. 185:4956–4972 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.